Peptide Selenocysteine Substitutions Reveal Direct Substrate-Enzyme Interactions at Auxiliary Clusters in Radical S-Adenosyl-l-methionine Maturases
- PMID: 37104670
- PMCID: PMC10177961
- DOI: 10.1021/jacs.3c00831
Peptide Selenocysteine Substitutions Reveal Direct Substrate-Enzyme Interactions at Auxiliary Clusters in Radical S-Adenosyl-l-methionine Maturases
Abstract
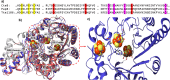
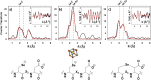
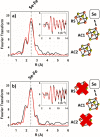
Radical S-adenosyl-l-methionine (SAM) enzymes leverage the properties of one or more iron- and sulfide-containing metallocenters to catalyze complex and radical-mediated transformations. By far the most populous superfamily of radical SAM enzymes are those that, in addition to a 4Fe-4S cluster that binds and activates the SAM cofactor, also bind one or more additional auxiliary clusters (ACs) of largely unknown catalytic significance. In this report we examine the role of ACs in two RS enzymes, PapB and Tte1186, that catalyze formation of thioether cross-links in ribosomally synthesized and post-translationally modified peptides (RiPPs). Both enzymes catalyze a sulfur-to-carbon cross-link in a reaction that entails H atom transfer from an unactivated C-H to initiate catalysis, followed by formation of a C-S bond to yield the thioether. We show that both enzymes tolerate substitution of SeCys instead of Cys at the cross-linking site, allowing the systems to be subjected to Se K-edge X-ray spectroscopy. The EXAFS data show a direct interaction with the Fe of one of the ACs in the Michaelis complex, which is replaced with a Se-C interaction under reducing conditions that lead to the product complex. Site-directed deletion of the clusters in Tte1186 provide evidence for the identity of the AC. The implications of these observations in the context of the mechanism of these thioether cross-linking enzymes are discussed.
Conflict of interest statement
The authors declare the following competing financial interest(s): Aspects of this work have been disclosed to the University of Utah, which holds patent interests in the findings.
Figures
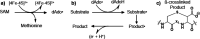

Similar articles
-
Biochemical and Spectroscopic Characterization of a Radical S-Adenosyl-L-methionine Enzyme Involved in the Formation of a Peptide Thioether Cross-Link.Biochemistry. 2016 Apr 12;55(14):2122-34. doi: 10.1021/acs.biochem.6b00145. Epub 2016 Apr 1. Biochemistry. 2016. PMID: 27007615 Free PMC article.
-
A Radical Clock Probe Uncouples H Atom Abstraction from Thioether Cross-Link Formation by the Radical S-Adenosyl-l-methionine Enzyme SkfB.Biochemistry. 2018 Aug 14;57(32):4816-4823. doi: 10.1021/acs.biochem.8b00537. Epub 2018 Jul 24. Biochemistry. 2018. PMID: 29965747 Free PMC article.
-
Intermolecular electron transfer in radical SAM enzymes as a new paradigm for reductive activation.J Biol Chem. 2023 Sep;299(9):105058. doi: 10.1016/j.jbc.2023.105058. Epub 2023 Jul 17. J Biol Chem. 2023. PMID: 37460016 Free PMC article.
-
Mechanism of Radical Initiation in the Radical S-Adenosyl-l-methionine Superfamily.Acc Chem Res. 2018 Nov 20;51(11):2611-2619. doi: 10.1021/acs.accounts.8b00356. Epub 2018 Oct 15. Acc Chem Res. 2018. PMID: 30346729 Free PMC article. Review.
-
Auxiliary iron-sulfur cofactors in radical SAM enzymes.Biochim Biophys Acta. 2015 Jun;1853(6):1316-34. doi: 10.1016/j.bbamcr.2015.01.002. Epub 2015 Jan 15. Biochim Biophys Acta. 2015. PMID: 25597998 Review.
Cited by
-
The Radical SAM Heme Synthase AhbD from Methanosarcina barkeri Contains Two Auxiliary [4Fe-4S] Clusters.Biomolecules. 2023 Aug 18;13(8):1268. doi: 10.3390/biom13081268. Biomolecules. 2023. PMID: 37627333 Free PMC article.
-
Ripping and stitching with copper.Nat Chem Biol. 2024 Apr;20(4):404-405. doi: 10.1038/s41589-024-01582-9. Nat Chem Biol. 2024. PMID: 38514883 No abstract available.
-
Benzylic Radical Stabilization Permits Ether Formation During Darobactin Biosynthesis.bioRxiv [Preprint]. 2023 Nov 29:2023.11.29.569256. doi: 10.1101/2023.11.29.569256. bioRxiv. 2023. PMID: 38076856 Free PMC article. Preprint.
-
Darobactin Substrate Engineering and Computation Show Radical Stability Governs Ether versus C-C Bond Formation.J Am Chem Soc. 2024 May 22;146(20):14328-14340. doi: 10.1021/jacs.4c03994. Epub 2024 May 10. J Am Chem Soc. 2024. PMID: 38728535 Free PMC article.
References
-
- Holliday G. L.; Akiva E.; Meng E. C.; Brown S. D.; Calhoun S.; Pieper U.; Sali A.; Booker S. J.; Babbitt P. C. Atlas of the Radical SAM Superfamily: Divergent Evolution of Function Using a ‘Plug and Play’ Domain. Methods in Enzymology 2018, 606, 1–71. 10.1016/bs.mie.2018.06.004. - DOI - PMC - PubMed
-
- Sofia H. J.; Chen G.; Hetzler B. G.; Reyes-Spindola J. F.; Miller N. E. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 2001, 29 (5), 1097–1106. 10.1093/nar/29.5.1097. - DOI - PMC - PubMed
-
- Walsby C. J.; Ortillo D.; Broderick W. E.; Broderick J. B.; Hoffman B. M. An Anchoring Role for FeS Cluster: Chelation of the Amino Acid Moiety of S-Adenosylmethionine to the Unique Iron Site of the [4Fe-4S] Cluster of Pyruvate Formate-Lyase Activating Enzyme. J. Am. Chem. Soc. 2002, 124 (38), 11270–11271. 10.1021/ja027078v. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

