Functional Diversity of HemN-like Proteins
- PMID: 37101745
- PMCID: PMC10114718
- DOI: 10.1021/acsbiomedchemau.1c00058
Functional Diversity of HemN-like Proteins
Abstract
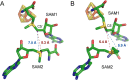
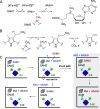
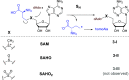
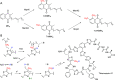
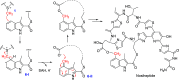
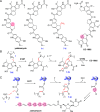
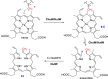
HemN is a radical S-adenosylmethionine (SAM) enzyme that catalyzes the anaerobic oxidative decarboxylation of coproporphyrinogen III to produce protoporphyrinogen IX, a key intermediate in heme biosynthesis. Proteins homologous to HemN (HemN-like proteins) are widespread in both prokaryotes and eukaryotes. Although these proteins are in most cases annotated as anaerobic coproporphyrinogen III oxidases (CPOs) in the public database, many of them are actually not CPOs but have diverse functions such as methyltransferases, cyclopropanases, heme chaperones, to name a few. This Perspective discusses the recent advances in the understanding of HemN-like proteins, and particular focus is placed on the diverse chemistries and functions of this growing protein family.
© 2022 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
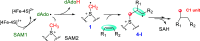
Similar articles
-
Revisiting the Mechanism of the Anaerobic Coproporphyrinogen III Oxidase HemN.Angew Chem Int Ed Engl. 2019 May 6;58(19):6235-6238. doi: 10.1002/anie.201814708. Epub 2019 Apr 1. Angew Chem Int Ed Engl. 2019. PMID: 30884058
-
Radical S-adenosylmethionine enzyme coproporphyrinogen III oxidase HemN: functional features of the [4Fe-4S] cluster and the two bound S-adenosyl-L-methionines.J Biol Chem. 2005 Aug 12;280(32):29038-46. doi: 10.1074/jbc.M501275200. Epub 2005 Jun 20. J Biol Chem. 2005. PMID: 15967800
-
The oxygen-independent coproporphyrinogen III oxidase HemN utilizes harderoporphyrinogen as a reaction intermediate during conversion of coproporphyrinogen III to protoporphyrinogen IX.Biol Chem. 2010 Jan;391(1):55-63. doi: 10.1515/BC.2010.006. Biol Chem. 2010. PMID: 19919179
-
Radical SAM Enzymes Involved in Tetrapyrrole Biosynthesis and Insertion.ACS Bio Med Chem Au. 2022 Feb 16;2(3):196-204. doi: 10.1021/acsbiomedchemau.1c00061. eCollection 2022 Jun 15. ACS Bio Med Chem Au. 2022. PMID: 37101575 Free PMC article. Review.
-
Recent advances in HemN-like radical S-adenosyl-l-methionine enzyme-catalyzed reactions.Nat Prod Rep. 2020 Jan 1;37(1):17-28. doi: 10.1039/c9np00032a. Epub 2019 Jul 10. Nat Prod Rep. 2020. PMID: 31290896 Review.
Cited by
-
Biosynthesis of coelulatin for the methylation of anthraquinone featuring HemN-like radical S-adenosyl-L-methionine enzyme.Front Microbiol. 2022 Nov 17;13:1040900. doi: 10.3389/fmicb.2022.1040900. eCollection 2022. Front Microbiol. 2022. PMID: 36466681 Free PMC article.
-
In Campylobacter jejuni, a new type of chaperone receives heme from ferrochelatase.Front Genet. 2023 Jun 21;14:1199357. doi: 10.3389/fgene.2023.1199357. eCollection 2023. Front Genet. 2023. PMID: 37415606 Free PMC article.
-
Twenty Years of Radical SAM! The Genesis of the Superfamily.ACS Bio Med Chem Au. 2022 Dec 5;2(6):538-547. doi: 10.1021/acsbiomedchemau.2c00078. eCollection 2022 Dec 21. ACS Bio Med Chem Au. 2022. PMID: 37101427 Free PMC article. No abstract available.
-
Identification of a poly-cyclopropylglycine-containing peptide via bioinformatic mapping of radical S-adenosylmethionine enzymes.J Biol Chem. 2022 May;298(5):101881. doi: 10.1016/j.jbc.2022.101881. Epub 2022 Mar 31. J Biol Chem. 2022. PMID: 35367210 Free PMC article.
-
Structure and Catalytic Mechanism of Radical SAM Methylases.Life (Basel). 2022 Oct 28;12(11):1732. doi: 10.3390/life12111732. Life (Basel). 2022. PMID: 36362886 Free PMC article. Review.
References
Publication types
LinkOut - more resources
Full Text Sources
