Thymidine phosphorylase facilitates retinoic acid inducible gene-I induced endothelial dysfunction
- PMID: 37100811
- PMCID: PMC10131517
- DOI: 10.1038/s41419-023-05821-0
Thymidine phosphorylase facilitates retinoic acid inducible gene-I induced endothelial dysfunction
Abstract
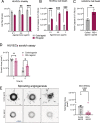
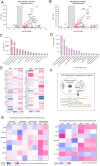
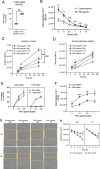
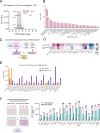
Activation of nucleic acid sensors in endothelial cells (ECs) has been shown to drive inflammation across pathologies including cancer, atherosclerosis and obesity. We previously showed that enhancing cytosolic DNA sensing by inhibiting three prime exonuclease 1 (TREX1) in ECs led to EC dysfunction and impaired angiogenesis. Here we show that activation of a cytosolic RNA sensor, Retinoic acid Induced Gene 1 (RIG-I) diminishes EC survival, angiogenesis and triggers tissue specific gene expression programs. We discovered a RIG-I dependent 7 gene signature that affects angiogenesis, inflammation and coagulation. Among these, we identified the thymidine phosphorylase TYMP as a key mediator of RIG-I induced EC dysfunction via its regulation of a subset of interferon stimulated genes. Our RIG-I induced gene signature was also conserved in the context of human diseases - in lung cancer vasculature and herpesvirus infection of lung endothelial cells. Pharmacological or genetic inhibition of TYMP rescues RIG-I induced EC death, migration arrest and restores sprouting angiogenesis. Interestingly, using RNAseq we identified a gene expression program that was RIG-I induced but TYMP dependent. Analysis of this dataset indicated that IRF1 and IRF8 dependent transcription is diminished in RIG-I activated cells when TYMP is inhibited. Functional RNAi screen of our TYMP dependent EC genes, we found that a group of 5 genes - Flot1, Ccl5, Vars2, Samd9l and Ube2l6 are critical for endothelial cell death mediated by RIG-I activation. Our observations identify mechanisms by which RIG-I drives EC dysfunction and define pathways that can be pharmacologically targeted to ameliorate RIG-I induced vascular inflammation.
© 2023. The Author(s).
Conflict of interest statement
The authors delare no competing interests.
Figures
Similar articles
-
Inhibition of Ongoing Influenza A Virus Replication Reveals Different Mechanisms of RIG-I Activation.J Virol. 2019 Mar 5;93(6):e02066-18. doi: 10.1128/JVI.02066-18. Print 2019 Mar 15. J Virol. 2019. PMID: 30602605 Free PMC article.
-
Cytosolic Sensors of Viral RNA Are Involved in the Production of Interleukin-6 via Toll-Like Receptor 3 Signaling in Human Glomerular Endothelial Cells.Kidney Blood Press Res. 2019;44(1):62-71. doi: 10.1159/000498837. Epub 2019 Feb 22. Kidney Blood Press Res. 2019. PMID: 30808838
-
RIG-I is a key antiviral interferon-stimulated gene against hepatitis E virus regardless of interferon production.Hepatology. 2017 Jun;65(6):1823-1839. doi: 10.1002/hep.29105. Epub 2017 May 3. Hepatology. 2017. PMID: 28195391
-
Endothelial Cells as a Key Cell Type for Innate Immunity: A Focused Review on RIG-I Signaling Pathway.Front Immunol. 2022 Jul 5;13:951614. doi: 10.3389/fimmu.2022.951614. eCollection 2022. Front Immunol. 2022. PMID: 35865527 Free PMC article. Review.
-
Thymidine phosphorylase: A potential new target for treating cardiovascular disease.Trends Cardiovasc Med. 2018 Apr;28(3):157-171. doi: 10.1016/j.tcm.2017.10.003. Epub 2017 Oct 20. Trends Cardiovasc Med. 2018. PMID: 29108898 Free PMC article. Review.
Cited by
-
A new gene signature for endothelial senescence identifies self-RNA sensing by retinoic acid-inducible gene I as a molecular facilitator of vascular aging.Aging Cell. 2024 Sep;23(9):e14240. doi: 10.1111/acel.14240. Epub 2024 Jun 21. Aging Cell. 2024. PMID: 39422883 Free PMC article.
-
Using machine learning model explanations to identify proteins related to severity of meibomian gland dysfunction.Sci Rep. 2023 Dec 22;13(1):22946. doi: 10.1038/s41598-023-50342-7. Sci Rep. 2023. PMID: 38135766 Free PMC article.
-
Lysosomal dysfunction and overload of nucleosides in thymidine phosphorylase deficiency of MNGIE.J Transl Med. 2024 May 13;22(1):449. doi: 10.1186/s12967-024-05275-8. J Transl Med. 2024. PMID: 38741129 Free PMC article.
-
Spatial multiomics atlas reveals smooth muscle phenotypic transformation and metabolic reprogramming in diabetic macroangiopathy.Cardiovasc Diabetol. 2024 Oct 12;23(1):358. doi: 10.1186/s12933-024-02458-x. Cardiovasc Diabetol. 2024. PMID: 39395983 Free PMC article.
-
Tumour evolution and microenvironment interactions in 2D and 3D space.Nature. 2024 Oct;634(8036):1178-1186. doi: 10.1038/s41586-024-08087-4. Epub 2024 Oct 30. Nature. 2024. PMID: 39478210 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

