High-Throughput Profiling of Candida auris Isolates Reveals Clade-Specific Metabolic Differences
- PMID: 37097196
- PMCID: PMC10269459
- DOI: 10.1128/spectrum.00498-23
High-Throughput Profiling of Candida auris Isolates Reveals Clade-Specific Metabolic Differences
Abstract
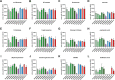
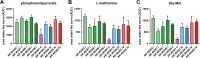
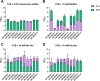
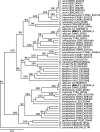
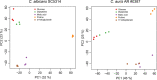
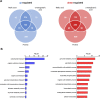
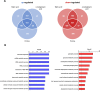
Candida auris, a multidrug-resistant human fungal pathogen that causes outbreaks of invasive infections, emerged as four distinct geographical clades. Previous studies identified genomic and proteomic differences in nutrient utilization on comparison to Candida albicans, suggesting that certain metabolic features may contribute to C. auris emergence. Since no high-throughput clade-specific metabolic characterization has been described yet, we performed a phenotypic screening of C. auris strains from all 4 clades on 664 nutrients, 120 chemicals, and 24 stressors. We identified common and clade- or strain-specific responses, including the preferred utilization of various dipeptides as nitrogen source and the inability of the clade II isolate AR 0381 to withstand chemical stress. Further analysis of the metabolic properties of C. auris isolates showed robust growth on intermediates of the tricarboxylic acid cycle, such as citrate and succinic and malic acids. However, there was reduced or no growth on pyruvate, lactic acid, or acetate, likely due to the lack of the monocarboxylic acid transporter Jen1, which is conserved in most pathogenic Candida species. Comparison of C. auris and C. albicans transcriptomes of cells grown on alternative carbon sources and dipeptides as a nitrogen source revealed common as well as species-unique responses. C. auris induced a significant number of genes with no ortholog in C. albicans, e.g., genes similar to the nicotinic acid transporter TNA1 (alternative carbon sources) and to the oligopeptide transporter (OPT) family (dipeptides). Thus, C. auris possesses unique metabolic features which could have contributed to its emergence as a pathogen. IMPORTANCE Four main clades of the emerging, multidrug-resistant human pathogen Candida auris have been identified, and they differ in their susceptibilities to antifungals and disinfectants. Moreover, clade- and strain-specific metabolic differences have been identified, but a comprehensive overview of nutritional characteristics and resistance to various stressors is missing. Here, we performed high-throughput phenotypic characterization of C. auris on various nutrients, stressors, and chemicals and obtained transcriptomes of cells grown on selected nutrients. The generated data sets identified multiple clade- and strain-specific phenotypes and induction of C. auris-specific metabolic genes, showing unique metabolic properties. The presented work provides a large amount of information for further investigations that could explain the role of metabolism in emergence and pathogenicity of this multidrug-resistant fungus.
Keywords: Candida auris; Jen2; carboxylic acids; dicarboxylic acids; dipeptide transport; dipeptides; metabolism; phenotypic profiling.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Emergence of the novel sixth Candida auris Clade VI in Bangladesh.Microbiol Spectr. 2024 Jul 2;12(7):e0354023. doi: 10.1128/spectrum.03540-23. Epub 2024 Jun 6. Microbiol Spectr. 2024. PMID: 38842332 Free PMC article.
-
Detection and characterisation of a sixth Candida auris clade in Singapore: a genomic and phenotypic study.Lancet Microbe. 2024 Sep;5(9):100878. doi: 10.1016/S2666-5247(24)00101-0. Epub 2024 Jul 13. Lancet Microbe. 2024. PMID: 39008997
-
ClaID: a Rapid Method of Clade-Level Identification of the Multidrug Resistant Human Fungal Pathogen Candida auris.Microbiol Spectr. 2022 Apr 27;10(2):e0063422. doi: 10.1128/spectrum.00634-22. Epub 2022 Mar 28. Microbiol Spectr. 2022. PMID: 35343775 Free PMC article.
-
A decade after the emergence of Candida auris: what do we know?Eur J Clin Microbiol Infect Dis. 2020 Sep;39(9):1617-1627. doi: 10.1007/s10096-020-03886-9. Epub 2020 Apr 15. Eur J Clin Microbiol Infect Dis. 2020. PMID: 32297040 Review.
-
Candida auris: Epidemiology, biology, antifungal resistance, and virulence.PLoS Pathog. 2020 Oct 22;16(10):e1008921. doi: 10.1371/journal.ppat.1008921. eCollection 2020 Oct. PLoS Pathog. 2020. PMID: 33091071 Free PMC article. Review.
Cited by
-
Sphingolipid diversity in Candida auris: unraveling interclade and drug resistance fingerprints.FEMS Yeast Res. 2024 Jan 9;24:foae008. doi: 10.1093/femsyr/foae008. FEMS Yeast Res. 2024. PMID: 38444195 Free PMC article.
-
Metabolic Patterns of Fluconazole Resistant and Susceptible Candida auris Clade V and I.J Fungi (Basel). 2024 Jul 25;10(8):518. doi: 10.3390/jof10080518. J Fungi (Basel). 2024. PMID: 39194844 Free PMC article.
-
What makes Candida auris pan-drug resistant? Integrative insights from genomic, transcriptomic, and phenomic analysis of clinical strains resistant to all four major classes of antifungal drugs.Antimicrob Agents Chemother. 2024 Oct 8;68(10):e0091124. doi: 10.1128/aac.00911-24. Epub 2024 Sep 19. Antimicrob Agents Chemother. 2024. PMID: 39297640
-
Metabolic homeostasis in fungal infections from the perspective of pathogens, immune cells, and whole-body systems.Microbiol Mol Biol Rev. 2024 Sep 26;88(3):e0017122. doi: 10.1128/mmbr.00171-22. Epub 2024 Sep 4. Microbiol Mol Biol Rev. 2024. PMID: 39230301 Review.
-
Tools and techniques to identify, study, and control Candida auris.PLoS Pathog. 2023 Oct 19;19(10):e1011698. doi: 10.1371/journal.ppat.1011698. eCollection 2023 Oct. PLoS Pathog. 2023. PMID: 37856418 Free PMC article. Review.
References
-
- Proctor DM, Dangana T, Sexton DJ, Fukuda C, Yelin RD, Stanley M, Bell PB, Baskaran S, Deming C, Chen Q, Conlan S, Park M, Mullikin J, Thomas J, Young A, Bouffard G, Barnabas B, Brooks S, Han J, Ho S-l, Kim J, Legaspi R, Maduro Q, Marfani H, Montemayor C, Riebow N, Schandler K, Schmidt B, Sison C, Stantripop M, Black S, Dekhtyar M, Masiello C, McDowell J, Thomas P, Vemulapalli M, Welsh RM, Vallabhaneni S, Chiller T, Forsberg K, Black SR, Pacilli M, Kong HH, Lin MY, Schoeny ME, Litvintseva AP, Segre JA, Hayden MK. 2021. Integrated genomic, epidemiologic investigation of Candida auris skin colonization in a skilled nursing facility. Nat Med 27:1401–1409. doi:10.1038/s41591-021-01383-w. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

