Reticulons promote formation of ER-derived double-membrane vesicles that facilitate SARS-CoV-2 replication
- PMID: 37093123
- PMCID: PMC10130743
- DOI: 10.1083/jcb.202203060
Reticulons promote formation of ER-derived double-membrane vesicles that facilitate SARS-CoV-2 replication
Abstract
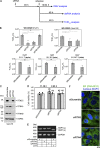
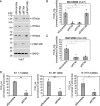
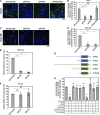
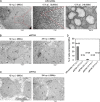
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), the etiologic agent for the global COVID-19 pandemic, triggers the formation of endoplasmic reticulum (ER)-derived replication organelles, including double-membrane vesicles (DMVs), in the host cell to support viral replication. Here, we clarify how SARS-CoV-2 hijacks host factors to construct the DMVs. We show that the ER morphogenic proteins reticulon-3 (RTN3) and RTN4 help drive DMV formation, enabling viral replication, which leads to productive infection. Different SARS-CoV-2 variants, including the delta variant, use the RTN-dependent pathway to promote infection. Mechanistically, our results reveal that the membrane-embedded reticulon homology domain (RHD) of the RTNs is sufficient to functionally support viral replication and physically engage NSP3 and NSP4, two viral non-structural membrane proteins known to induce DMV formation. Our findings thus identify the ER morphogenic RTN3 and RTN4 membrane proteins as host factors that help promote the biogenesis of SARS-CoV-2-induced DMVs, which can act as viral replication platforms.
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures


Comment in
-
RTN3 and RTN4: Architects of SARS-CoV-2 replication organelles.J Cell Biol. 2023 Jul 3;222(7):e202306020. doi: 10.1083/jcb.202306020. Epub 2023 Jun 15. J Cell Biol. 2023. PMID: 37318453 Free PMC article.
Similar articles
-
Expression and Cleavage of Middle East Respiratory Syndrome Coronavirus nsp3-4 Polyprotein Induce the Formation of Double-Membrane Vesicles That Mimic Those Associated with Coronaviral RNA Replication.mBio. 2017 Nov 21;8(6):e01658-17. doi: 10.1128/mBio.01658-17. mBio. 2017. PMID: 29162711 Free PMC article.
-
Oligomeric assembly of the C-terminal and transmembrane region of SARS-CoV-2 nsp3.J Virol. 2024 Apr 16;98(4):e0157523. doi: 10.1128/jvi.01575-23. Epub 2024 Mar 14. J Virol. 2024. PMID: 38483167 Free PMC article.
-
RTN3 and RTN4: Architects of SARS-CoV-2 replication organelles.J Cell Biol. 2023 Jul 3;222(7):e202306020. doi: 10.1083/jcb.202306020. Epub 2023 Jun 15. J Cell Biol. 2023. PMID: 37318453 Free PMC article.
-
The double-membrane vesicle (DMV): a virus-induced organelle dedicated to the replication of SARS-CoV-2 and other positive-sense single-stranded RNA viruses.Cell Mol Life Sci. 2022 Jul 16;79(8):425. doi: 10.1007/s00018-022-04469-x. Cell Mol Life Sci. 2022. PMID: 35841484 Free PMC article. Review.
-
The cell edit: Looking at and beyond non-structural proteins to understand membrane rearrangement in coronaviruses.Arch Biochem Biophys. 2024 Feb;752:109856. doi: 10.1016/j.abb.2023.109856. Epub 2023 Dec 15. Arch Biochem Biophys. 2024. PMID: 38104958 Review.
Cited by
-
Identification of Three Novel Linear B-Cell Epitopes in Non-Structural Protein 3 of Porcine Epidemic Diarrhea Virus Using Monoclonal Antibodies.Viruses. 2024 Mar 9;16(3):424. doi: 10.3390/v16030424. Viruses. 2024. PMID: 38543789 Free PMC article.
-
What do we know about the function of SARS-CoV-2 proteins?Front Immunol. 2023 Sep 18;14:1249607. doi: 10.3389/fimmu.2023.1249607. eCollection 2023. Front Immunol. 2023. PMID: 37790934 Free PMC article. Review.
-
Mutations in nonstructural proteins essential for pathogenicity in SARS-CoV-2-infected mice.J Virol. 2024 Jul 23;98(7):e0058424. doi: 10.1128/jvi.00584-24. Epub 2024 Jun 18. J Virol. 2024. PMID: 38888344 Free PMC article.
-
The glycoprotein quality control factor Malectin promotes coronavirus replication and viral protein biogenesis.bioRxiv [Preprint]. 2024 Jul 16:2024.06.02.597051. doi: 10.1101/2024.06.02.597051. bioRxiv. 2024. PMID: 38895409 Free PMC article. Preprint.
-
Identification of host dependency factors involved in SARS-CoV-2 replication organelle formation through proteomics and ultrastructural analysis.J Virol. 2023 Nov 30;97(11):e0087823. doi: 10.1128/jvi.00878-23. Epub 2023 Oct 31. J Virol. 2023. PMID: 37905840 Free PMC article.
References
-
- Al-Mulla, H.M., Turrell L., Smith N.M., Payne L., Baliji S., Züst R., Thiel V., Baker S.C., Siddell S.G., and Neuman B.W.. 2014. Competitive fitness in coronaviruses is not correlated with size or number of double-membrane vesicles under reduced-temperature growth conditions. MBio. 5:e01107–e01113. 10.1128/mBio.01107-13 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

