Identification of the cell cycle characteristics of non-small cell lung cancer and its relationship with tumor immune microenvironment, cell death pathways, and metabolic reprogramming
- PMID: 37091844
- PMCID: PMC10117961
- DOI: 10.3389/fendo.2023.1147366
Identification of the cell cycle characteristics of non-small cell lung cancer and its relationship with tumor immune microenvironment, cell death pathways, and metabolic reprogramming
Abstract
Background: The genes related to the cell cycle progression could be considered the key factors in human cancers. However, the genes involved in cell cycle regulation in non-small cell lung cancer (NSCLC) have not yet been reported. Therefore, it is necessary to evaluate the genes related to the cell cycle in all types of cancers, especially NSCLC.
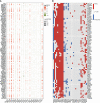
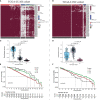
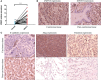
Methods: This study constituted the first pan-cancer landscape of cell cycle signaling. Cluster analysis based on cell cycle signaling was conducted to identify the potential molecular heterogeneity of NSCLC. Further, the discrepancies in the tumor immune microenvironment, metabolic remodeling, and cell death among the three clusters were investigated. Immunohistochemistry was performed to validate the protein levels of the ZWINT gene and examine its relationship with the clinical characteristics. Bioinformatics analyses and experimental validation of the ZWINT gene were also conducted.
Results: First, pan-cancer analysis provided an overview of cell cycle signaling and highlighted its crucial role in cancer. A majority of cell cycle regulators play risk roles in lung adenocarcinoma (LUAD); however, some cell cycle genes play protective roles in lung squamous cell carcinoma (LUSC). Cluster analysis revealed three potential subtypes for patients with NSCLC. LUAD patients with high cell cycle activities were associated with worse prognosis; while, LUSC patients with high cell cycle activities were associated with a longer survival time. Moreover, the above three subtypes of NSCLC exhibited distinct immune microenvironments, metabolic remodeling, and cell death pathways. ZWINT, a member of the cell signaling pathway, was observed to be significantly associated with the prognosis of LUAD patients. A series of experiments verified the higher expression levels of ZWINT in NSCLC compared to those in paracancerous tissues. The activation of epithelial-mesenchymal transition (EMT) induced by ZWINT might be responsible for tumor progression.
Conclusion: This study revealed the regulatory function of the cell cycle genes in NSCLC, and the molecular classification based on cell cycle-associated genes could evaluate the different prognoses of patients with NSCLC. ZWINT expression was found to be significantly upregulated in NSCLC tissues, which might promote tumor progression via activation of the EMT pathway.
Keywords: cell cycle; cell death pathways; metabolic reprogramming; non-small cell lung cancer; pan-cancer analysis; tumor immune microenvironment.
Copyright © 2023 Cao, Xiao, Zhang and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







Similar articles
-
Insights into the heterogeneity of the tumor microenvironment in lung adenocarcinoma and squamous carcinoma through single-cell transcriptomic analysis: Implications for distinct immunotherapy outcomes.J Gene Med. 2024 Jun;26(6):e3694. doi: 10.1002/jgm.3694. J Gene Med. 2024. PMID: 38847309
-
ZNF143 Expression is Associated with COPD and Tumor Microenvironment in Non-Small Cell Lung Cancer.Int J Chron Obstruct Pulmon Dis. 2022 Apr 2;17:685-700. doi: 10.2147/COPD.S352392. eCollection 2022. Int J Chron Obstruct Pulmon Dis. 2022. PMID: 35400998 Free PMC article.
-
The heterogeneous immune landscape between lung adenocarcinoma and squamous carcinoma revealed by single-cell RNA sequencing.Signal Transduct Target Ther. 2022 Aug 26;7(1):289. doi: 10.1038/s41392-022-01130-8. Signal Transduct Target Ther. 2022. PMID: 36008393 Free PMC article.
-
The Effect of GLUT1 on the Survival Rate and Immune Cell Infiltration of Lung Adenocarcinoma and Squamous Cell Carcinoma: A Meta and Bioinformatics Analysis.Anticancer Agents Med Chem. 2022;22(2):223-238. doi: 10.2174/1871520621666210708115406. Anticancer Agents Med Chem. 2022. PMID: 34238200 Review.
-
Cellular Senescence in Non-Small Cell Lung Cancer.Front Biosci (Landmark Ed). 2023 Dec 28;28(12):357. doi: 10.31083/j.fbl2812357. Front Biosci (Landmark Ed). 2023. PMID: 38179738 Review.
Cited by
-
[Relationship between GTSE1 and Cell Cycle and Potential Regulatory Mechanisms in Lung Cancer Cells].Zhongguo Fei Ai Za Zhi. 2024 Jun 20;27(6):451-458. doi: 10.3779/j.issn.1009-3419.2024.106.13. Zhongguo Fei Ai Za Zhi. 2024. PMID: 39026496 Free PMC article. Review. Chinese.
-
Unveiling Potential Targeted Therapeutic Opportunities for Co-Overexpressed Targeting Protein for Xklp2 and Aurora-A Kinase in Lung Adenocarcinoma.Mol Biotechnol. 2024 Oct;66(10):2792-2803. doi: 10.1007/s12033-023-00879-9. Epub 2023 Sep 28. Mol Biotechnol. 2024. PMID: 37768502 Free PMC article.
-
The crosstalk role of CDKN2A between tumor progression and cuproptosis resistance in colorectal cancer.Aging (Albany NY). 2024 Jun 17;16(12):10512-10538. doi: 10.18632/aging.205945. Epub 2024 Jun 17. Aging (Albany NY). 2024. PMID: 38888512 Free PMC article.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

