Molecular characterization of coxsackievirus B5 from the sputum of pneumonia children patients of Kunming, Southwest China
- PMID: 37076847
- PMCID: PMC10116704
- DOI: 10.1186/s12985-023-02019-w
Molecular characterization of coxsackievirus B5 from the sputum of pneumonia children patients of Kunming, Southwest China
Abstract
Background: CVB5 can cause respiratory infections. However, the molecular epidemiological information about CVB5 in respiratory tract samples is still limited. Here, we report five cases in which CVB5 was detected in sputum sample of pneumonia children patients from Kunming, Southwest China.
Methods: CVB5 isolates were obtained from sputum samples of patients with pneumonia. Whole-genome sequencing of CVB5 isolates was performed using segmented PCR, and phylogenetic, mutation and recombination analysis. The effect of mutations in the VP1 protein on hydration were analyzed by Protscale. The tertiary models of VP1 proteins were established by Colabfold, and the effect of mutations in VP1 protein on volume modifications and binding affinity were analyzed by Pymol software and PROVEAN.
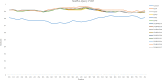
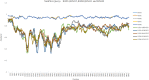
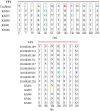
Results: A total of five CVB5 complete genome sequences were obtained. No obvious homologous recombination signals comparing with other coxsackie B viruses were observed in the five isolates. Phylogenetic analysis showed that the five CVB5 sputum isolates were from an independent branch in genogroup E. Due to the mutation, the structure and spatial of the VP1 protein N-terminus have changed significantly. Comparing to the Faulkner (CVB5 prototype strain), PROVEAN revealed three deleterious substitutions: Y75F, N166T (KM35), T140I (KM41). The last two of the three deleterious substitutions significantly increased the hydrophobicity of the residues.
Conclusions: We unexpectedly found five cases of CVB5 infection instead of rhinoviruses infection during our routine surveillance of rhinoviruses in respiratory tract samples. All five patients were hospitalized with pneumonia symptoms and were not tested for enterovirus during their hospitalization. This report suggests that enterovirus surveillance in patients with respiratory symptoms should be strengthened.
Keywords: Analysis; CVB5; Epidemiology; Pneumonia.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


Similar articles
-
Recombination analysis of coxsackievirus B5 genogroup C.Arch Virol. 2018 Feb;163(2):539-544. doi: 10.1007/s00705-017-3608-6. Epub 2017 Nov 13. Arch Virol. 2018. PMID: 29134337
-
[Complete genome sequence of Coxsackievirus B5 (CVB5/09) strain isolated in China and its phylogenetic analysis].Bing Du Xue Bao. 2010 Jul;26(4):283-9. Bing Du Xue Bao. 2010. PMID: 20836381 Chinese.
-
Analysis of Coxsackievirus B5 Infections in the Central Nervous System in Brazil: Insights into Molecular Epidemiology and Genetic Diversity.Viruses. 2022 Apr 26;14(5):899. doi: 10.3390/v14050899. Viruses. 2022. PMID: 35632640 Free PMC article.
-
Molecular Epidemiology Reveals the Co-Circulation of Two Genotypes of Coxsackievirus B5 in China.Viruses. 2022 Nov 30;14(12):2693. doi: 10.3390/v14122693. Viruses. 2022. PMID: 36560696 Free PMC article.
-
Clinical and molecular epidemiologic features of enterovirus D68 infection in children with acute lower respiratory tract infection in China.Arch Virol. 2023 Jul 15;168(8):206. doi: 10.1007/s00705-023-05823-5. Arch Virol. 2023. PMID: 37453955 Review.
References
-
- Andréoletti L, Renois F, Jacques J, Lévêque N. Entérovirus non poliomyélitiques et pathologies respiratoires [Human enteroviruses and respiratory infections] Med Sci. 2009;25(11):921–930. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

