Perfect duet: Dual recombinases improve genetic resolution
- PMID: 37060165
- PMCID: PMC10212704
- DOI: 10.1111/cpr.13446
Perfect duet: Dual recombinases improve genetic resolution
Abstract
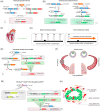
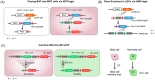
As a powerful genetic tool, site-specific recombinases (SSRs) have been widely used in genomic manipulation to elucidate cell fate plasticity in vivo, advancing research in stem cell and regeneration medicine. However, the low resolution of conventional single-recombinase-mediated lineage tracing strategies, which rely heavily on the specificity of one marker gene, has led to controversial conclusions in many scientific questions. Therefore, different SSRs systems are combined to improve the accuracy of lineage tracing. Here we review the recent advances in dual-recombinase-mediated genetic approaches, including the development of novel genetic recombination technologies and their applications in cell differentiation, proliferation, and genetic manipulation. In comparison with the single-recombinase system, we also discuss the advantages of dual-genetic strategies in solving scientific issues as well as their technical limitations.
© 2023 The Authors. Cell Proliferation published by Beijing Institute for Stem Cell and Regenerative Medicine and John Wiley & Sons Ltd.
Conflict of interest statement
No conflict of interest.
Figures


Similar articles
-
Dual recombinases-based genetic lineage tracing for stem cell research with enhanced precision.Sci China Life Sci. 2021 Dec;64(12):2060-2072. doi: 10.1007/s11427-020-1889-9. Epub 2021 Apr 9. Sci China Life Sci. 2021. PMID: 33847909 Review.
-
Triple-cell lineage tracing by a dual reporter on a single allele.J Biol Chem. 2020 Jan 17;295(3):690-700. doi: 10.1074/jbc.RA119.011349. Epub 2019 Nov 26. J Biol Chem. 2020. PMID: 31771978 Free PMC article.
-
Genetic lineage tracing with multiple DNA recombinases: A user's guide for conducting more precise cell fate mapping studies.J Biol Chem. 2020 May 8;295(19):6413-6424. doi: 10.1074/jbc.REV120.011631. Epub 2020 Mar 25. J Biol Chem. 2020. PMID: 32213599 Free PMC article. Review.
-
Enhancing the precision of genetic lineage tracing using dual recombinases.Nat Med. 2017 Dec;23(12):1488-1498. doi: 10.1038/nm.4437. Epub 2017 Nov 13. Nat Med. 2017. PMID: 29131159 Free PMC article.
-
Harnessing orthogonal recombinases to decipher cell fate with enhanced precision.Trends Cell Biol. 2022 Apr;32(4):324-337. doi: 10.1016/j.tcb.2021.09.007. Epub 2021 Oct 15. Trends Cell Biol. 2022. PMID: 34657762 Review.
References
-
- Blanpain C, Simons BD. Unravelling stem cell dynamics by lineage tracing. Nat Rev Mol Cell Biol. 2013;14(8):489‐502. - PubMed
-
- VanHorn S, Morris SA. Next‐generation lineage tracing and fate mapping to interrogate development. Dev Cell. 2021;56(1):7‐21. - PubMed
-
- Nagy A. Cre recombinase: the universal reagent for genome tailoring. Genesis. 2000;26(2):99‐109. - PubMed
-
- Anastassiadis K, Fu J, Patsch C, et al. Dre recombinase, like Cre, is a highly efficient site‐specific recombinase in E. coli, mammalian cells and mice. Dis Model Mech. 2009;2(9–10):508‐515. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

