Intact Transition Epitope Mapping-Serological Inspection by Epitope EXtraction (ITEM-SIX)
- PMID: 37049857
- PMCID: PMC10096252
- DOI: 10.3390/molecules28073092
Intact Transition Epitope Mapping-Serological Inspection by Epitope EXtraction (ITEM-SIX)
Abstract
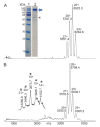
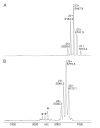
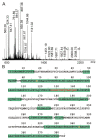
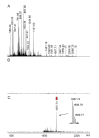
Precision medicine requests accurate serological inspections to precisely stratify patients for targeted treatment. Intact transition epitope mapping analysis proved surrogate seroconversion of a model organism's serum when spiked with a monoclonal murine anti-Ovalbumin antibody (mAb) with epitope resolution. Isolation of the IgG fraction from blood serum applied two consecutive protein precipitation steps followed by ultrafiltration and resulted in an ESI-MS analysis-ready IgG preparation. For epitope mapping by epitope extraction, the Ovalbumin antigen was digested with trypsin. After desalting, the peptide mixture was added to the ESI-MS-ready IgG preparation from mAb-spiked serum and the solution was incubated to form an immune complex between the Ovalbumin-derived epitope peptide and the anti-Ovalbumin mAb. Then, the entire mixture of proteins and peptides was directly electrosprayed. Sorting of ions in the mass spectrometer's gas phase, dissociation of the immune complex ions by collision-induced dissociation, and recording of the epitope peptide ion that had been released from the immune complex proved the presence of the anti-Ovalbumin mAb in serum. Mass determination of the complex-released epitope peptide ion with isotope resolution is highly accurate, guaranteeing high specificity of this novel analysis approach, which is termed Intact Transition Epitope Mapping-Serological Inspections by Epitope EXtraction (ITEM-SIX).
Keywords: blood serum; epitope mapping; immune complex analysis; nanoESI mass spectrometry; surrogate seroconversion.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Intact Transition Epitope Mapping - Thermodynamic Weak-force Order (ITEM - TWO).J Proteomics. 2020 Feb 10;212:103572. doi: 10.1016/j.jprot.2019.103572. Epub 2019 Nov 1. J Proteomics. 2020. PMID: 31683061
-
Intact Transition Epitope Mapping - Targeted High-Energy Rupture of Extracted Epitopes (ITEM-THREE).Mol Cell Proteomics. 2019 Aug;18(8):1543-1555. doi: 10.1074/mcp.RA119.001429. Epub 2019 May 30. Mol Cell Proteomics. 2019. PMID: 31147491 Free PMC article.
-
Intact Transition Epitope Mapping (ITEM).J Am Soc Mass Spectrom. 2017 Aug;28(8):1612-1622. doi: 10.1007/s13361-017-1654-7. Epub 2017 Jun 14. J Am Soc Mass Spectrom. 2017. PMID: 28616748
-
Immuno-Affinity Mass Spectrometry: A Novel Approaches with Biomedical Relevance.Adv Exp Med Biol. 2019;1140:377-388. doi: 10.1007/978-3-030-15950-4_21. Adv Exp Med Biol. 2019. PMID: 31347059 Review.
-
Mass spectrometric epitope mapping.Mass Spectrom Rev. 2018 Mar;37(2):229-241. doi: 10.1002/mas.21516. Epub 2016 Jul 12. Mass Spectrom Rev. 2018. PMID: 27403762 Review.
Cited by
-
Mass Spectrometric ITEM-ONE and ITEM-TWO Analyses Confirm and Refine an Assembled Epitope of an Anti-Pertuzumab Affimer.Biomolecules. 2023 Dec 24;14(1):24. doi: 10.3390/biom14010024. Biomolecules. 2023. PMID: 38254624 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

