NAGS, CPS1, and SLC25A13 (Citrin) at the Crossroads of Arginine and Pyrimidines Metabolism in Tumor Cells
- PMID: 37047726
- PMCID: PMC10094985
- DOI: 10.3390/ijms24076754
NAGS, CPS1, and SLC25A13 (Citrin) at the Crossroads of Arginine and Pyrimidines Metabolism in Tumor Cells
Abstract
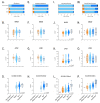
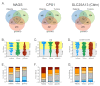
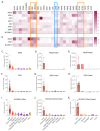
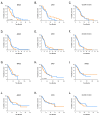
Urea cycle enzymes and transporters collectively convert ammonia into urea in the liver. Aberrant overexpression of carbamylphosphate synthetase 1 (CPS1) and SLC25A13 (citrin) genes has been associated with faster proliferation of tumor cells due to metabolic reprogramming that increases the activity of the CAD complex and pyrimidine biosynthesis. N-acetylglutamate (NAG), produced by NAG synthase (NAGS), is an essential activator of CPS1. Although NAGS is expressed in lung cancer derived cell lines, expression of the NAGS gene and its product was not evaluated in tumors with aberrant expression of CPS1 and citrin. We used data mining approaches to identify tumor types that exhibit aberrant overexpression of NAGS, CPS1, and citrin genes, and evaluated factors that may contribute to increased expression of the three genes and their products in tumors. Median expression of NAGS, CPS1, and citrin mRNA was higher in glioblastoma multiforme (GBM), glioma, and stomach adenocarcinoma (STAD) samples compared to the matched normal tissue. Median expression of CPS1 and citrin mRNA was higher in the lung adenocarcinoma (LUAD) sample while expression of NAGS mRNA did not differ. High NAGS expression was associated with an unfavorable outcome in patients with glioblastoma and GBM. Low NAGS expression was associated with an unfavorable outcome in patients with LUAD. Patterns of DNase hypersensitive sites and histone modifications in the upstream regulatory regions of NAGS, CPS1, and citrin genes were similar in liver tissue, lung tissue, and A549 lung adenocarcinoma cells despite different expression levels of the three genes in the liver and lung. Citrin gene copy numbers correlated with its mRNA expression in glioblastoma, GBM, LUAD, and STAD samples. There was little overlap between NAGS, CPS1, and citrin sequence variants found in patients with respective deficiencies, tumor samples, and individuals without known rare genetic diseases. The correlation between NAGS, CPS1, and citrin mRNA expression in the individual glioblastoma, GBM, LUAD, and STAD samples was very weak. These results suggest that the increased cytoplasmic supply of either carbamylphosphate, produced by CPS1, or aspartate may be sufficient to promote tumorigenesis, as well as the need for an alternative explanation of CPS1 activity in the absence of NAGS expression and NAG.
Keywords: CAD complex; N-acetylglutamate synthase; carbamylphosphate synthetase 1; citrin; pyrimidine biosynthesis; tumor; tumorigenesis; urea cycle.
Conflict of interest statement
Ljubica Caldovic’s work was supported by the Recordati Rare Diseases, Inc. that manufactures and sells NCG as Carbaglu® (carglumic acid), which is used for the treatment of
Figures

Similar articles
-
Noncoding sequence variants define a novel regulatory element in the first intron of the N-acetylglutamate synthase gene.Hum Mutat. 2021 Dec;42(12):1624-1636. doi: 10.1002/humu.24281. Epub 2021 Sep 24. Hum Mutat. 2021. PMID: 34510628 Free PMC article.
-
N-acetylglutamate synthase deficiency: an insight into the genetics, epidemiology, pathophysiology, and treatment.Appl Clin Genet. 2011 Aug 24;4:127-35. doi: 10.2147/TACG.S12702. Print 2011. Appl Clin Genet. 2011. PMID: 23776373 Free PMC article.
-
Expression pattern and biochemical properties of zebrafish N-acetylglutamate synthase.PLoS One. 2014 Jan 22;9(1):e85597. doi: 10.1371/journal.pone.0085597. eCollection 2014. PLoS One. 2014. PMID: 24465614 Free PMC article.
-
The N-Acetylglutamate Synthase Family: Structures, Function and Mechanisms.Int J Mol Sci. 2015 Jun 9;16(6):13004-22. doi: 10.3390/ijms160613004. Int J Mol Sci. 2015. PMID: 26068232 Free PMC article. Review.
-
N-acetylglutamate synthase: structure, function and defects.Mol Genet Metab. 2010;100 Suppl 1(Suppl 1):S13-9. doi: 10.1016/j.ymgme.2010.02.018. Epub 2010 Feb 26. Mol Genet Metab. 2010. PMID: 20303810 Free PMC article. Review.
Cited by
-
Construction of a novel lipid drop-mitochondria-associated genetic profile for predicting the survival and prognosis of lung adenocarcinoma.Discov Oncol. 2024 Nov 17;15(1):668. doi: 10.1007/s12672-024-01526-8. Discov Oncol. 2024. PMID: 39551861 Free PMC article.
References
-
- Jin G., Reitman Z.J., Duncan C.G., Spasojevic I., Gooden D.M., Rasheed B.A., Yang R., Lopez G.Y., He Y., McLendon R.E., et al. Disruption of wild-type IDH1 suppresses D-2-hydroxyglutarate production in IDH1-mutated gliomas. Cancer Res. 2013;73:496–501. doi: 10.1158/0008-5472.CAN-12-2852. - DOI - PMC - PubMed
-
- Hartmann C., Meyer J., Balss J., Capper D., Mueller W., Christians A., Felsberg J., Wolter M., Mawrin C., Wick W., et al. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: A study of 1010 diffuse gliomas. Acta Neuropathol. 2009;118:469–474. doi: 10.1007/s00401-009-0561-9. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

