Deregulated Transcription and Proteostasis in Adult mapt Knockout Mouse
- PMID: 37047532
- PMCID: PMC10095510
- DOI: 10.3390/ijms24076559
Deregulated Transcription and Proteostasis in Adult mapt Knockout Mouse
Abstract
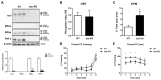
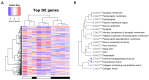
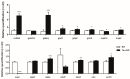
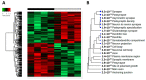
Transcriptomics and phosphoproteomics were carried out in the cerebral cortex of B6.Cg-Mapttm1(EGFP)Klt (tau knockout: tau-KO) and wild-type (WT) 12 month-old mice to learn about the effects of tau ablation. Compared with WT mice, tau-KO mice displayed reduced anxiety-like behavior and lower fear expression induced by aversive conditioning, whereas recognition memory remained unaltered. Cortical transcriptomic analysis revealed 69 downregulated and 105 upregulated genes in tau-KO mice, corresponding to synaptic structures, neuron cytoskeleton and transport, and extracellular matrix components. RT-qPCR validated increased mRNA levels of col6a4, gabrq, gad1, grm5, grip2, map2, rab8a, tubb3, wnt16, and an absence of map1a in tau-KO mice compared with WT mice. A few proteins were assessed with Western blotting to compare mRNA expression with corresponding protein levels. Map1a mRNA and protein levels decreased. However, β-tubulin III and GAD1 protein levels were reduced in tau-KO mice. Cortical phosphoproteomics revealed 121 hypophosphorylated and 98 hyperphosphorylated proteins in tau-KO mice. Deregulated phosphoproteins were categorized into cytoskeletal (n = 45) and membrane proteins, including proteins of the synapses and vesicles, myelin proteins, and proteins linked to membrane transport and ion channels (n = 84), proteins related to DNA and RNA metabolism (n = 36), proteins connected to the ubiquitin-proteasome system (UPS) (n = 7), proteins with kinase or phosphatase activity (n = 21), and 22 other proteins related to variegated pathways such as metabolic pathways, growth factors, or mitochondrial function or structure. The present observations reveal a complex altered brain transcriptome and phosphoproteome in tau-KO mice with only mild behavioral alterations.
Keywords: cytoskeleton; phosphoproteomics; synapse; tau-KO; transcriptomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Familial globular glial tauopathy linked to MAPT mutations: molecular neuropathology and seeding capacity of a prototypical mixed neuronal and glial tauopathy.Acta Neuropathol. 2020 Apr;139(4):735-771. doi: 10.1007/s00401-019-02122-9. Epub 2020 Jan 6. Acta Neuropathol. 2020. PMID: 31907603 Free PMC article.
-
Loss of tau and Fyn reduces compensatory effects of MAP2 for tau and reveals a Fyn-independent effect of tau on calcium.J Neurosci Res. 2019 Nov;97(11):1393-1413. doi: 10.1002/jnr.24517. Epub 2019 Aug 26. J Neurosci Res. 2019. PMID: 31452242 Free PMC article.
-
Decrease of neuronal FKBP4/FKBP52 modulates perinuclear lysosomal positioning and MAPT/Tau behavior during MAPT/Tau-induced proteotoxic stress.Autophagy. 2021 Nov;17(11):3491-3510. doi: 10.1080/15548627.2021.1875611. Epub 2021 Jan 25. Autophagy. 2021. PMID: 33459145 Free PMC article.
-
Cellular and molecular modifier pathways in tauopathies: the big picture from screening invertebrate models.J Neurochem. 2016 Apr;137(1):12-25. doi: 10.1111/jnc.13532. Epub 2016 Feb 11. J Neurochem. 2016. PMID: 26756400 Review.
-
Disrupted ubiquitin proteasome system underlying tau accumulation in Alzheimer's disease.Neurobiol Aging. 2021 Mar;99:79-85. doi: 10.1016/j.neurobiolaging.2020.11.015. Epub 2020 Dec 8. Neurobiol Aging. 2021. PMID: 33422896 Review.
Cited by
-
The disease-causing tau V337M mutation induces tau hypophosphorylation and perturbs axon morphology pathways.bioRxiv [Preprint]. 2024 Jun 6:2024.06.04.597496. doi: 10.1101/2024.06.04.597496. bioRxiv. 2024. PMID: 38895329 Free PMC article. Preprint.
-
Alzheimer's disease: an axonal injury disease?Front Aging Neurosci. 2023 Oct 19;15:1264448. doi: 10.3389/fnagi.2023.1264448. eCollection 2023. Front Aging Neurosci. 2023. PMID: 37927337 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

