SIR telomere silencing depends on nuclear envelope lipids and modulates sensitivity to a lysolipid
- PMID: 37042812
- PMCID: PMC10103788
- DOI: 10.1083/jcb.202206061
SIR telomere silencing depends on nuclear envelope lipids and modulates sensitivity to a lysolipid
Abstract
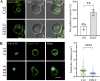
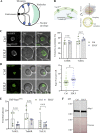
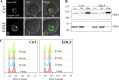
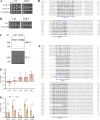
The nuclear envelope (NE) is important in maintaining genome organization. The role of lipids in communication between the NE and telomere regulation was investigated, including how changes in lipid composition impact gene expression and overall nuclear architecture. Yeast was treated with the non-metabolizable lysophosphatidylcholine analog edelfosine, known to accumulate at the perinuclear ER. Edelfosine induced NE deformation and disrupted telomere clustering but not anchoring. Additionally, the association of Sir4 at telomeres decreased. RNA-seq analysis showed altered expression of Sir-dependent genes located at sub-telomeric (0-10 kb) regions, consistent with Sir4 dispersion. Transcriptomic analysis revealed that two lipid metabolic circuits were activated in response to edelfosine, one mediated by the membrane sensing transcription factors, Spt23/Mga2, and the other by a transcriptional repressor, Opi1. Activation of these transcriptional programs resulted in higher levels of unsaturated fatty acids and the formation of nuclear lipid droplets. Interestingly, cells lacking Sir proteins displayed resistance to unsaturated-fatty acids and edelfosine, and this phenotype was connected to Rap1.
© 2023 Sosa Ponce et al.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures




Similar articles
-
The functional importance of telomere clustering: global changes in gene expression result from SIR factor dispersion.Genome Res. 2009 Apr;19(4):611-25. doi: 10.1101/gr.083881.108. Epub 2009 Jan 29. Genome Res. 2009. PMID: 19179643 Free PMC article.
-
Subtelomeric factors antagonize telomere anchoring and Tel1-independent telomere length regulation.EMBO J. 2006 Feb 22;25(4):857-67. doi: 10.1038/sj.emboj.7600976. Epub 2006 Feb 9. EMBO J. 2006. PMID: 16467853 Free PMC article.
-
Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1.Genes Dev. 1994 Oct 1;8(19):2257-69. doi: 10.1101/gad.8.19.2257. Genes Dev. 1994. PMID: 7958893
-
The function of telomere clustering in yeast: the circe effect.Cold Spring Harb Symp Quant Biol. 2004;69:327-37. doi: 10.1101/sqb.2004.69.327. Cold Spring Harb Symp Quant Biol. 2004. PMID: 16117665 Review. No abstract available.
-
Multiple pathways tether telomeres and silent chromatin at the nuclear periphery: functional implications for sir-mediated repression.Novartis Found Symp. 2005;264:140-56; discussion 156-65, 227-30. Novartis Found Symp. 2005. PMID: 15773752 Review.
Cited by
-
Dual activities of a silencing information regulator complex in yeast transcriptional regulation and DNA-damage response.mLife. 2024 May 15;3(2):207-218. doi: 10.1002/mlf2.12108. eCollection 2024 Jun. mLife. 2024. PMID: 38948145 Free PMC article. Review.
References
-
- Ames, B.N. 1966. Assay of inorganic phosphate, total phosphate and phosphatases. Methods Enzymol. 8:115–118. 10.1016/0076-6879(66)08014-5 - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

