Comprehensive investigation of the expression profiles of common long noncoding RNAs during microglial activation
- PMID: 37037460
- PMCID: PMC10085744
- DOI: 10.5808/gi.22061
Comprehensive investigation of the expression profiles of common long noncoding RNAs during microglial activation
Abstract
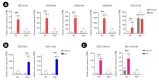
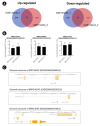
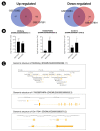
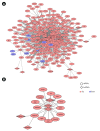
Microglia, similar to peripheral macrophages, are the primary immune cells of the central nervous system (CNS). Microglia exist in the resting state in the healthy CNS, but can be activated and polarized into either M1 or M2 subtypes for immune defense and the maintenance of CNS homeostasis by multiple stimuli. Several long noncoding RNAs (lncRNAs) mediate human inflammatory diseases and neuropathologies by regulating their target genes. However, the function of common lncRNAs that contribute to microglial activation remains unclear. Thus, we used bioinformatic approaches to identify common lncRNAs involved in microglial activation in vitro. Our study identified several lncRNAs as common regulators of microglial activation. We identified 283 common mRNAs and 53 common lncRNAs during mouse M1 microglial activation processes, whereas 26 common mRNAs and five common lncRNAs were identified during mouse M2 microglial activation processes. A total of 648 common mRNAs and 274 common lncRNAs were identified during the activation of human M1 microglia. In addition, we identified 1,920 common co-expressed pairs in mouse M1 activation processes and 25 common co-expressed pairs in mouse M2 activation processes. Our study provides a comprehensive understanding of common lncRNA expression profiles in microglial activation processes in vitro. The list of common lncRNAs identified in this study provides novel evidence and clues regarding the molecular mechanisms underlying microglial activation.
Keywords: M1 microglia activation; M2 microglia activation; lnc-miRHGs; long noncoding RNA; microglia.
Conflict of interest statement
No potential conflict of interest relevant to this article was reported.
Figures

Similar articles
-
The potential roles of m6A modification in regulating the inflammatory response in microglia.J Neuroinflammation. 2021 Jul 5;18(1):149. doi: 10.1186/s12974-021-02205-z. J Neuroinflammation. 2021. PMID: 34225746 Free PMC article.
-
Long Noncoding RNA Profiling from Fasciola Gigantica Excretory/Secretory Product-Induced M2 to M1 Macrophage Polarization.Cell Physiol Biochem. 2018;47(2):505-522. doi: 10.1159/000489984. Epub 2018 May 22. Cell Physiol Biochem. 2018. PMID: 29794463
-
Comprehensive analysis of mRNAs, lncRNAs and circRNAs in the early phase of microglial activation.Exp Ther Med. 2021 Dec;22(6):1460. doi: 10.3892/etm.2021.10895. Epub 2021 Oct 20. Exp Ther Med. 2021. PMID: 34737800 Free PMC article.
-
Role of microglial m1/m2 polarization in relapse and remission of psychiatric disorders and diseases.Pharmaceuticals (Basel). 2014 Nov 25;7(12):1028-48. doi: 10.3390/ph7121028. Pharmaceuticals (Basel). 2014. PMID: 25429645 Free PMC article. Review.
-
One locus with two roles: microRNA-independent functions of microRNA-host-gene locus-encoded long noncoding RNAs.Wiley Interdiscip Rev RNA. 2021 May;12(3):e1625. doi: 10.1002/wrna.1625. Epub 2020 Sep 17. Wiley Interdiscip Rev RNA. 2021. PMID: 32945142 Free PMC article. Review.
References
-
- Ling EA, Ng YK, Wu CH, Kaur C. Microglia: its development and role as a neuropathology sensor. Prog Brain Res. 2001;132:61–79. - PubMed
-
- Dickson DW, Lee SC, Mattiace LA, Yen SH, Brosnan C. Microglia and cytokines in neurological disease, with special reference to AIDS and Alzheimer's disease. Glia. 1993;7:75–83. - PubMed
-
- Cacabelos R, Alvarez XA, Fernandez-Novoa L, Franco A, Mangues R, Pellicer A, et al. Brain interleukin-1 beta in Alzheimer's disease and vascular dementia. Methods Find Exp Clin Pharmacol. 1994;16:141–151. - PubMed
-
- Bauer J, Konig G, Strauss S, Jonas U, Ganter U, Weidemann A, et al. In-vitro matured human macrophages express Alzheimer's beta A4-amyloid precursor protein indicating synthesis in microglial cells. FEBS Lett. 1991;282:335–340. - PubMed
LinkOut - more resources
Full Text Sources

