Skeletal muscle transcriptomics dissects the pathogenesis of Friedreich's ataxia
- PMID: 37027192
- PMCID: PMC10281753
- DOI: 10.1093/hmg/ddad051
Skeletal muscle transcriptomics dissects the pathogenesis of Friedreich's ataxia
Abstract
Objective: In Friedreich's ataxia (FRDA), the most affected tissues are not accessible to sampling and available transcriptomic findings originate from blood-derived cells and animal models. Herein, we aimed at dissecting for the first time the pathophysiology of FRDA by means of RNA-sequencing in an affected tissue sampled in vivo.
Methods: Skeletal muscle biopsies were collected from seven FRDA patients before and after treatment with recombinant human Erythropoietin (rhuEPO) within a clinical trial. Total RNA extraction, 3'-mRNA library preparation and sequencing were performed according to standard procedures. We tested for differential gene expression with DESeq2 and performed gene set enrichment analysis with respect to control subjects.
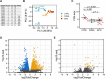
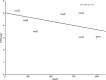
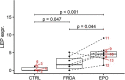
Results: FRDA transcriptomes showed 1873 genes differentially expressed from controls. Two main signatures emerged: (1) a global downregulation of the mitochondrial transcriptome as well as of ribosome/translational machinery and (2) an upregulation of genes related to transcription and chromatin regulation, especially of repressor terms. Downregulation of the mitochondrial transcriptome was more profound than previously shown in other cellular systems. Furthermore, we observed in FRDA patients a marked upregulation of leptin, the master regulator of energy homeostasis. RhuEPO treatment further enhanced leptin expression.
Interpretation: Our findings reflect a double hit in the pathophysiology of FRDA: a transcriptional/translational issue and a profound mitochondrial failure downstream. Leptin upregulation in the skeletal muscle in FRDA may represent a compensatory mechanism of mitochondrial dysfunction, which is amenable to pharmacological boosting. Skeletal muscle transcriptomics is a valuable biomarker to monitor therapeutic interventions in FRDA.
© The Author(s) 2022. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures


Similar articles
-
Skeletal Muscle Involvement in Friedreich Ataxia.Int J Mol Sci. 2024 Sep 13;25(18):9915. doi: 10.3390/ijms25189915. Int J Mol Sci. 2024. PMID: 39337401 Free PMC article. Review.
-
Comprehensive analysis of gene expression patterns in Friedreich's ataxia fibroblasts by RNA sequencing reveals altered levels of protein synthesis factors and solute carriers.Dis Model Mech. 2017 Nov 1;10(11):1353-1369. doi: 10.1242/dmm.030536. Dis Model Mech. 2017. PMID: 29125828 Free PMC article.
-
Skeletal muscle involvement in friedreich ataxia and potential effects of recombinant human erythropoietin administration on muscle regeneration and neovascularization.J Neuropathol Exp Neurol. 2012 Aug;71(8):708-15. doi: 10.1097/NEN.0b013e31825fed76. J Neuropathol Exp Neurol. 2012. PMID: 22805773
-
Bioenergetics of the calf muscle in Friedreich ataxia patients measured by 31P-MRS before and after treatment with recombinant human erythropoietin.PLoS One. 2013 Jul 29;8(7):e69229. doi: 10.1371/journal.pone.0069229. Print 2013. PLoS One. 2013. PMID: 23922695 Free PMC article. Clinical Trial.
-
Mitochondrial dysfunction in Friedreich's ataxia: from pathogenesis to treatment perspectives.Free Radic Res. 2002 Apr;36(4):461-6. doi: 10.1080/10715760290021324. Free Radic Res. 2002. PMID: 12069111 Review.
Cited by
-
Skeletal Muscle Involvement in Friedreich Ataxia.Int J Mol Sci. 2024 Sep 13;25(18):9915. doi: 10.3390/ijms25189915. Int J Mol Sci. 2024. PMID: 39337401 Free PMC article. Review.
-
Skeletal muscle proteome analysis underpins multifaceted mitochondrial dysfunction in Friedreich's ataxia.Front Neurosci. 2023 Oct 31;17:1289027. doi: 10.3389/fnins.2023.1289027. eCollection 2023. Front Neurosci. 2023. PMID: 38027498 Free PMC article.
-
Differential Gene Expression in Late-Onset Friedreich Ataxia: A Comparative Transcriptomic Analysis Between Symptomatic and Asymptomatic Sisters.Int J Mol Sci. 2024 Oct 29;25(21):11615. doi: 10.3390/ijms252111615. Int J Mol Sci. 2024. PMID: 39519164 Free PMC article.
References
-
- Vankan, P. (2013) Prevalence gradients of Friedreich's ataxia and R1b haplotype in Europe co-localize, suggesting a common Palaeolithic origin in the Franco-Cantabrian ice age refuge. J. Neurochem., 126, 11–20. - PubMed
-
- Saveliev, A., Everett, C., Sharpe, T., Webster, Z. and Festenstein, R. (2003) DNA triplet repeats mediate heterochromatin-protein-1-sensitive variegated gene silencing. Nature, 422, 909–913. - PubMed
-
- Rodden, L.N., Chutake, Y.K., Gilliam, K., Lam, C., Soragni, E., Hauser, L., Gilliam, M., Wiley, G., Anderson, M.P., Gottesfeld, J.M., Lynch, D.R. and Bidichandani, S.I. (2021) Methylated and unmethylated epialleles support variegated epigenetic silencing in Friedreich ataxia. Hum. Mol. Genet., 29, 3818–3829. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

