Recent advances in therapeutic CRISPR-Cas9 genome editing: mechanisms and applications
- PMID: 37027099
- PMCID: PMC10080534
- DOI: 10.1186/s43556-023-00115-5
Recent advances in therapeutic CRISPR-Cas9 genome editing: mechanisms and applications
Abstract
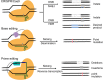
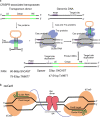
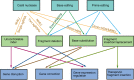
Recently, clustered regularly interspaced palindromic repeats (CRISPR)-Cas9 derived editing tools had significantly improved our ability to make desired changes in the genome. Wild-type Cas9 protein recognizes the target genomic loci and induced local double strand breaks (DSBs) in the guidance of small RNA molecule. In mammalian cells, the DSBs are mainly repaired by endogenous non-homologous end joining (NHEJ) pathway, which is error prone and results in the formation of indels. The indels can be harnessed to interrupt gene coding sequences or regulation elements. The DSBs can also be fixed by homology directed repair (HDR) pathway to introduce desired changes, such as base substitution and fragment insertion, when proper donor templates are provided, albeit in a less efficient manner. Besides making DSBs, Cas9 protein can be mutated to serve as a DNA binding platform to recruit functional modulators to the target loci, performing local transcriptional regulation, epigenetic remolding, base editing or prime editing. These Cas9 derived editing tools, especially base editors and prime editors, can introduce precise changes into the target loci at a single-base resolution and in an efficient and irreversible manner. Such features make these editing tools very promising for therapeutic applications. This review focuses on the evolution and mechanisms of CRISPR-Cas9 derived editing tools and their applications in the field of gene therapy.
Keywords: Base editing; CRISPR/Cas9; Gene Delivery; Gene therapy; Prime editing.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
A high-efficiency and versatile CRISPR/Cas9-mediated HDR-based biallelic editing system.J Zhejiang Univ Sci B. 2022 Feb 15;23(2):141-152. doi: 10.1631/jzus.B2100196. J Zhejiang Univ Sci B. 2022. PMID: 35187887 Free PMC article. English.
-
CRISPR-Cas9 fusion to dominant-negative 53BP1 enhances HDR and inhibits NHEJ specifically at Cas9 target sites.Nat Commun. 2019 Jun 28;10(1):2866. doi: 10.1038/s41467-019-10735-7. Nat Commun. 2019. PMID: 31253785 Free PMC article.
-
Advance trends in targeting homology-directed repair for accurate gene editing: An inclusive review of small molecules and modified CRISPR-Cas9 systems.Bioimpacts. 2022;12(4):371-391. doi: 10.34172/bi.2022.23871. Epub 2022 Jun 22. Bioimpacts. 2022. PMID: 35975201 Free PMC article. Review.
-
CRISPR-Cas nucleases and base editors for plant genome editing.aBIOTECH. 2019 Nov 30;1(1):74-87. doi: 10.1007/s42994-019-00010-0. eCollection 2020 Jan. aBIOTECH. 2019. PMID: 36305010 Free PMC article. Review.
-
Gene Editing With TALEN and CRISPR/Cas in Rice.Prog Mol Biol Transl Sci. 2017;149:81-98. doi: 10.1016/bs.pmbts.2017.04.006. Epub 2017 May 24. Prog Mol Biol Transl Sci. 2017. PMID: 28712502 Review.
Cited by
-
Advances in targeting cancer epigenetics using CRISPR-dCas9 technology: A comprehensive review and future prospects.Funct Integr Genomics. 2024 Sep 18;24(5):164. doi: 10.1007/s10142-024-01455-3. Funct Integr Genomics. 2024. PMID: 39292321 Review.
-
Off-the-Shelf Cord-Blood Mesenchymal Stromal Cells: Production, Quality Control, and Clinical Use.Cells. 2024 Jun 19;13(12):1066. doi: 10.3390/cells13121066. Cells. 2024. PMID: 38920694 Free PMC article.
-
What's in a cure: designing a broad-spectrum HIV gene therapy.Curr Opin HIV AIDS. 2024 May 1;19(3):150-156. doi: 10.1097/COH.0000000000000846. Epub 2024 Mar 1. Curr Opin HIV AIDS. 2024. PMID: 38547339 Free PMC article. Review.
-
Engineering CRISPR/Cas9 therapeutics for cancer precision medicine.Front Genet. 2024 Apr 25;15:1309175. doi: 10.3389/fgene.2024.1309175. eCollection 2024. Front Genet. 2024. PMID: 38725484 Free PMC article. Review.
-
A Lipid Nanoparticle-Based Method for the Generation of Liver-Specific Knockout Mice.Int J Mol Sci. 2023 Sep 19;24(18):14299. doi: 10.3390/ijms241814299. Int J Mol Sci. 2023. PMID: 37762602 Free PMC article.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
