LPS-aggregating proteins GBP1 and GBP2 are each sufficient to enhance caspase-4 activation both in cellulo and in vitro
- PMID: 37023136
- PMCID: PMC10104521
- DOI: 10.1073/pnas.2216028120
LPS-aggregating proteins GBP1 and GBP2 are each sufficient to enhance caspase-4 activation both in cellulo and in vitro
Abstract
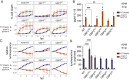
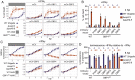
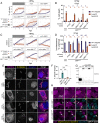
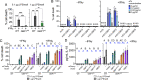
The gamma-interferon (IFNγ)-inducible guanylate-binding proteins (GBPs) promote host defense against gram-negative cytosolic bacteria in part through the induction of an inflammatory cell death pathway called pyroptosis. To activate pyroptosis, GBPs facilitate sensing of the gram-negative bacterial outer membrane component lipopolysaccharide (LPS) by the noncanonical caspase-4 inflammasome. There are seven human GBP paralogs, and it is unclear how each GBP contributes to LPS sensing and pyroptosis induction. GBP1 forms a multimeric microcapsule on the surface of cytosolic bacteria through direct interactions with LPS. The GBP1 microcapsule recruits caspase-4 to bacteria, a process deemed essential for caspase-4 activation. In contrast to GBP1, closely related paralog GBP2 is unable to bind bacteria on its own but requires GBP1 for direct bacterial binding. Unexpectedly, we find that GBP2 overexpression can restore gram-negative-induced pyroptosis in GBP1KO cells, without GBP2 binding to the bacterial surface. A mutant of GBP1 that lacks the triple arginine motif required for microcapsule formation also rescues pyroptosis in GBP1KO cells, showing that binding to bacteria is dispensable for GBPs to promote pyroptosis. Instead, we find that GBP2, like GBP1, directly binds and aggregates "free" LPS through protein polymerization. We demonstrate that supplementation of either recombinant polymerized GBP1 or GBP2 to an in vitro reaction is sufficient to enhance LPS-induced caspase-4 activation. This provides a revised mechanistic framework for noncanonical inflammasome activation where GBP1 or GBP2 assembles cytosol-contaminating LPS into a protein-LPS interface for caspase-4 activation as part of a coordinated host response to gram-negative bacterial infections.
Keywords: cell-autonomous immunity; guanylate-binding proteins; inflammasome; interferon-stimulated genes; lipopolysaccharide.
Conflict of interest statement
The authors declare no competing interest.
Figures


Similar articles
-
Shigella IpaH9.8 limits GBP1-dependent LPS release from intracytosolic bacteria to suppress caspase-4 activation.Proc Natl Acad Sci U S A. 2023 Apr 11;120(15):e2218469120. doi: 10.1073/pnas.2218469120. Epub 2023 Apr 4. Proc Natl Acad Sci U S A. 2023. PMID: 37014865 Free PMC article.
-
Human GBP1 binds LPS to initiate assembly of a caspase-4 activating platform on cytosolic bacteria.Nat Commun. 2020 Jun 24;11(1):3276. doi: 10.1038/s41467-020-16889-z. Nat Commun. 2020. PMID: 32581219 Free PMC article.
-
Inflammasome Activation by Bacterial Outer Membrane Vesicles Requires Guanylate Binding Proteins.mBio. 2017 Oct 3;8(5):e01188-17. doi: 10.1128/mBio.01188-17. mBio. 2017. PMID: 28974614 Free PMC article.
-
Human guanylate binding proteins: nanomachines orchestrating host defense.FEBS J. 2021 Oct;288(20):5826-5849. doi: 10.1111/febs.15662. Epub 2021 Jan 12. FEBS J. 2021. PMID: 33314740 Free PMC article. Review.
-
Interferon-inducible guanylate-binding proteins at the interface of cell-autonomous immunity and inflammasome activation.J Leukoc Biol. 2017 Jan;101(1):143-150. doi: 10.1189/jlb.4MR0516-223R. Epub 2016 Jul 14. J Leukoc Biol. 2017. PMID: 27418355 Free PMC article. Review.
Cited by
-
Pyroptosis in defense against intracellular bacteria.Semin Immunol. 2023 Sep;69:101805. doi: 10.1016/j.smim.2023.101805. Epub 2023 Jul 8. Semin Immunol. 2023. PMID: 37429234 Free PMC article. Review.
-
Deciphering the molecular nexus between Omicron infection and acute kidney injury: a bioinformatics approach.Front Mol Biosci. 2024 Jul 4;11:1340611. doi: 10.3389/fmolb.2024.1340611. eCollection 2024. Front Mol Biosci. 2024. PMID: 39027131 Free PMC article.
-
Caspase-11 drives macrophage hyperinflammation in models of Polg-related mitochondrial disease.bioRxiv [Preprint]. 2024 May 25:2024.05.11.593693. doi: 10.1101/2024.05.11.593693. bioRxiv. 2024. PMID: 38798587 Free PMC article. Preprint.
-
Newcastle disease virus promotes pyroptosis in medulloblastoma cells by regulating interferon-gamma-mediated guanylate-binding protein 1 expression and activating caspase-4.Cytojournal. 2024 Oct 30;21:39. doi: 10.25259/Cytojournal_39_2024. eCollection 2024. Cytojournal. 2024. PMID: 39563668 Free PMC article.
-
The role of galectins in the regulation of autophagy and inflammasome in host immunity.Semin Immunopathol. 2024 Jul 23;46(3-4):6. doi: 10.1007/s00281-024-01018-5. Semin Immunopathol. 2024. PMID: 39042263 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

