An updated atlas of antibody evasion by SARS-CoV-2 Omicron sub-variants including BQ.1.1 and XBB
- PMID: 37019110
- PMCID: PMC10027947
- DOI: 10.1016/j.xcrm.2023.100991
An updated atlas of antibody evasion by SARS-CoV-2 Omicron sub-variants including BQ.1.1 and XBB
Abstract
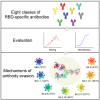
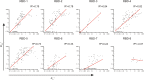
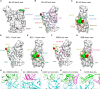
Emerging Omicron sub-variants are causing global concerns, and their immune evasion should be monitored continuously. We previously evaluated the escape of Omicron BA.1, BA.1.1, BA.2, and BA.3 from an atlas of 50 monoclonal antibodies (mAbs), covering seven epitope classes of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) receptor-binding domain (RBD). Here, we update the atlas of totally 77 mAbs against emerging sub-variants including BQ.1.1 and XBB and find that BA.4/5, BQ.1.1, and XBB display further evasion. Besides, investigation into the correlation of binding and neutralization of mAbs reveals the important role of antigenic conformation in mAb functioning. Moreover, the complex structures of BA.2 RBD/BD-604/S304 and BA.4/5 RBD/BD-604/S304/S309 further elucidate the molecular mechanism of antibody evasion by these sub-variants. By focusing on the identified broadly potent mAbs, we find a general hotspot epitope on the RBD, which could guide the design of vaccines and calls for new broad-spectrum countermeasures against COVID-19.
Keywords: BA.2.13; BA.2.75; BA.4; BA.5; BQ.1.1; Omicron BA.2.12.1; SARS-CoV-2; XBB; human neutralizing antibodies; immune escape.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures

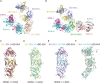


Similar articles
-
Defining a de novo non-RBM antibody as RBD-8 and its synergistic rescue of immune-evaded antibodies to neutralize Omicron SARS-CoV-2.Proc Natl Acad Sci U S A. 2023 Dec 26;120(52):e2314193120. doi: 10.1073/pnas.2314193120. Epub 2023 Dec 18. Proc Natl Acad Sci U S A. 2023. PMID: 38109549 Free PMC article.
-
A pan-sarbecovirus vaccine based on RBD of SARS-CoV-2 original strain elicits potent neutralizing antibodies against XBB in non-human primates.Proc Natl Acad Sci U S A. 2023 Mar 14;120(11):e2221713120. doi: 10.1073/pnas.2221713120. Epub 2023 Mar 10. Proc Natl Acad Sci U S A. 2023. PMID: 36897979 Free PMC article.
-
Atlas of currently available human neutralizing antibodies against SARS-CoV-2 and escape by Omicron sub-variants BA.1/BA.1.1/BA.2/BA.3.Immunity. 2022 Aug 9;55(8):1501-1514.e3. doi: 10.1016/j.immuni.2022.06.005. Epub 2022 Jun 15. Immunity. 2022. PMID: 35777362 Free PMC article.
-
Immune evasion of neutralizing antibodies by SARS-CoV-2 Omicron.Cytokine Growth Factor Rev. 2023 Apr;70:13-25. doi: 10.1016/j.cytogfr.2023.03.001. Epub 2023 Mar 5. Cytokine Growth Factor Rev. 2023. PMID: 36948931 Free PMC article. Review.
-
Antibody evasion associated with the RBD significant mutations in several emerging SARS-CoV-2 variants and its subvariants.Drug Resist Updat. 2023 Nov;71:101008. doi: 10.1016/j.drup.2023.101008. Epub 2023 Sep 22. Drug Resist Updat. 2023. PMID: 37757651 Review.
Cited by
-
Immune Evasion, Infectivity, and Fusogenicity of SARS-CoV-2 Omicron BA.2.86 and FLip Variants.bioRxiv [Preprint]. 2023 Sep 12:2023.09.11.557206. doi: 10.1101/2023.09.11.557206. bioRxiv. 2023. Update in: Cell. 2024 Feb 1;187(3):585-595.e6. doi: 10.1016/j.cell.2023.12.026 PMID: 37745517 Free PMC article. Updated. Preprint.
-
Key mechanistic features of the trade-off between antibody escape and host cell binding in the SARS-CoV-2 Omicron variant spike proteins.EMBO J. 2024 Apr;43(8):1484-1498. doi: 10.1038/s44318-024-00062-z. Epub 2024 Mar 11. EMBO J. 2024. PMID: 38467833 Free PMC article.
-
Immune evasion and membrane fusion of SARS-CoV-2 XBB subvariants EG.5.1 and XBB.2.3.Emerg Microbes Infect. 2023 Dec;12(2):2270069. doi: 10.1080/22221751.2023.2270069. Epub 2023 Oct 26. Emerg Microbes Infect. 2023. PMID: 37819267 Free PMC article.
-
Cross-species recognition of bat coronavirus RsYN04 and cross-reaction of SARS-CoV-2 antibodies against the virus.Zool Res. 2023 Nov 18;44(6):1015-1025. doi: 10.24272/j.issn.2095-8137.2023.187. Zool Res. 2023. PMID: 37804113 Free PMC article.
-
Exploring the ability of the MD+FoldX method to predict SARS-CoV-2 antibody escape mutations using large-scale data.Sci Rep. 2024 Oct 4;14(1):23122. doi: 10.1038/s41598-024-72491-z. Sci Rep. 2024. PMID: 39366988 Free PMC article.
References
-
- Saxena S.K., Kumar S., Ansari S., Paweska J.T., Maurya V.K., Tripathi A.K., Abdel-Moneim A.S. Transmission dynamics and mutational prevalence of the novel Severe acute respiratory syndrome coronavirus-2 Omicron Variant of Concern. J. Med. Virol. 2022;94:2160–2166. doi: 10.1002/jmv.27611. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

