Targeting Enterococcus faecalis HMG-CoA reductase with a non-statin inhibitor
- PMID: 37012403
- PMCID: PMC10070635
- DOI: 10.1038/s42003-023-04639-y
Targeting Enterococcus faecalis HMG-CoA reductase with a non-statin inhibitor
Abstract
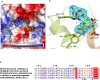
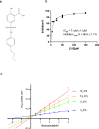
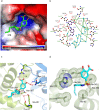
HMG-CoA reductase (HMGR), a rate-limiting enzyme of the mevalonate pathway in Gram-positive pathogenic bacteria, is an attractive target for development of novel antibiotics. In this study, we report the crystal structures of HMGR from Enterococcus faecalis (efHMGR) in the apo and liganded forms, highlighting several unique features of this enzyme. Statins, which inhibit the human enzyme with nanomolar affinity, perform poorly against the bacterial HMGR homologs. We also report a potent competitive inhibitor (Chembridge2 ID 7828315 or compound 315) of the efHMGR enzyme identified by a high-throughput, in-vitro screening. The X-ray crystal structure of efHMGR in complex with 315 was determined to 1.27 Å resolution revealing that the inhibitor occupies the mevalonate-binding site and interacts with several key active site residues conserved among bacterial homologs. Importantly, 315 does not inhibit the human HMGR. Our identification of a selective, non-statin inhibitor of bacterial HMG-CoA reductases will be instrumental in lead optimization and development of novel antibacterial drug candidates.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Enterococcus faecalis acetoacetyl-coenzyme A thiolase/3-hydroxy-3-methylglutaryl-coenzyme A reductase, a dual-function protein of isopentenyl diphosphate biosynthesis.J Bacteriol. 2002 Apr;184(8):2116-22. doi: 10.1128/JB.184.8.2116-2122.2002. J Bacteriol. 2002. PMID: 11914342 Free PMC article.
-
Structural mechanism for statin inhibition of HMG-CoA reductase.Science. 2001 May 11;292(5519):1160-4. doi: 10.1126/science.1059344. Science. 2001. PMID: 11349148
-
A fungal tolerance trait and selective inhibitors proffer HMG-CoA reductase as a herbicide mode-of-action.Nat Commun. 2022 Sep 22;13(1):5563. doi: 10.1038/s41467-022-33185-0. Nat Commun. 2022. PMID: 36137996 Free PMC article.
-
Structural mechanism for statin inhibition of 3-hydroxy-3-methylglutaryl coenzyme A reductase.Am Heart J. 2002 Dec;144(6 Suppl):S27-32. doi: 10.1067/mhj.2002.130300. Am Heart J. 2002. PMID: 12486413 Review.
-
The structure of the catalytic portion of human HMG-CoA reductase.Biochim Biophys Acta. 2000 Dec 15;1529(1-3):9-18. doi: 10.1016/s1388-1981(00)00134-7. Biochim Biophys Acta. 2000. PMID: 11111074 Review.
Cited by
-
Targeting the autophagy-miRNA axis in prostate cancer: toward novel diagnostic and therapeutic strategies.Naunyn Schmiedebergs Arch Pharmacol. 2024 Oct;397(10):7421-7437. doi: 10.1007/s00210-024-03153-0. Epub 2024 May 18. Naunyn Schmiedebergs Arch Pharmacol. 2024. PMID: 38761210 Review.
-
pH-dependent reaction triggering in PmHMGR crystals for time-resolved crystallography.Biophys J. 2024 Mar 5;123(5):622-637. doi: 10.1016/j.bpj.2024.02.003. Epub 2024 Feb 6. Biophys J. 2024. PMID: 38327055
-
Repositioning of HMG-CoA Reductase Inhibitors as Adjuvants in the Modulation of Efflux Pump-Mediated Bacterial and Tumor Resistance.Antibiotics (Basel). 2023 Sep 20;12(9):1468. doi: 10.3390/antibiotics12091468. Antibiotics (Basel). 2023. PMID: 37760764 Free PMC article. Review.
-
The Effects of Tocotrienol on Gut Microbiota: A Scoping Review.Life (Basel). 2023 Sep 7;13(9):1882. doi: 10.3390/life13091882. Life (Basel). 2023. PMID: 37763286 Free PMC article. Review.
References
-
- Johnson EA, Schroeder WA. Microbial carotenoids. Adv. Biochem. Eng. Biotechnol. 1996;53:119–178. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

