Geographically weighted linear combination test for gene-set analysis of a continuous spatial phenotype as applied to intratumor heterogeneity
- PMID: 36998245
- PMCID: PMC10044624
- DOI: 10.3389/fcell.2023.1065586
Geographically weighted linear combination test for gene-set analysis of a continuous spatial phenotype as applied to intratumor heterogeneity
Abstract
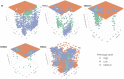
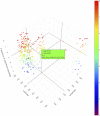
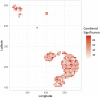
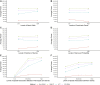
Background: The impact of gene-sets on a spatial phenotype is not necessarily uniform across different locations of cancer tissue. This study introduces a computational platform, GWLCT, for combining gene set analysis with spatial data modeling to provide a new statistical test for location-specific association of phenotypes and molecular pathways in spatial single-cell RNA-seq data collected from an input tumor sample. Methods: The main advantage of GWLCT consists of an analysis beyond global significance, allowing the association between the gene-set and the phenotype to vary across the tumor space. At each location, the most significant linear combination is found using a geographically weighted shrunken covariance matrix and kernel function. Whether a fixed or adaptive bandwidth is determined based on a cross-validation cross procedure. Our proposed method is compared to the global version of linear combination test (LCT), bulk and random-forest based gene-set enrichment analyses using data created by the Visium Spatial Gene Expression technique on an invasive breast cancer tissue sample, as well as 144 different simulation scenarios. Results: In an illustrative example, the new geographically weighted linear combination test, GWLCT, identifies the cancer hallmark gene-sets that are significantly associated at each location with the five spatially continuous phenotypic contexts in the tumors defined by different well-known markers of cancer-associated fibroblasts. Scan statistics revealed clustering in the number of significant gene-sets. A spatial heatmap of combined significance over all selected gene-sets is also produced. Extensive simulation studies demonstrate that our proposed approach outperforms other methods in the considered scenarios, especially when the spatial association increases. Conclusion: Our proposed approach considers the spatial covariance of gene expression to detect the most significant gene-sets affecting a continuous phenotype. It reveals spatially detailed information in tissue space and can thus play a key role in understanding the contextual heterogeneity of cancer cells.
Keywords: Spatial single cell analysis; cancer-associated fibroblast; gene-set analysis; geographically weighted regression; intratumor heterogeneity; linear combination test.
Copyright © 2023 Amini, Hajihosseini, Pyne and Dinu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Folic acid supplementation and malaria susceptibility and severity among people taking antifolate antimalarial drugs in endemic areas.Cochrane Database Syst Rev. 2022 Feb 1;2(2022):CD014217. doi: 10.1002/14651858.CD014217. Cochrane Database Syst Rev. 2022. PMID: 36321557 Free PMC article.
-
Linear combination test for gene set analysis of a continuous phenotype.BMC Bioinformatics. 2013 Jul 1;14:212. doi: 10.1186/1471-2105-14-212. BMC Bioinformatics. 2013. PMID: 23815123 Free PMC article.
-
Modeling crash spatial heterogeneity: random parameter versus geographically weighting.Accid Anal Prev. 2015 Feb;75:16-25. doi: 10.1016/j.aap.2014.10.020. Epub 2014 Nov 16. Accid Anal Prev. 2015. PMID: 25460087
-
Determination of the best geographic weighted function and estimation of spatio temporal model - Geographically weighted panel regression using weighted least square.MethodsX. 2024 Feb 6;12:102605. doi: 10.1016/j.mex.2024.102605. eCollection 2024 Jun. MethodsX. 2024. PMID: 38357634 Free PMC article. Review.
-
Translational Metabolomics of Head Injury: Exploring Dysfunctional Cerebral Metabolism with Ex Vivo NMR Spectroscopy-Based Metabolite Quantification.In: Kobeissy FH, editor. Brain Neurotrauma: Molecular, Neuropsychological, and Rehabilitation Aspects. Boca Raton (FL): CRC Press/Taylor & Francis; 2015. Chapter 25. In: Kobeissy FH, editor. Brain Neurotrauma: Molecular, Neuropsychological, and Rehabilitation Aspects. Boca Raton (FL): CRC Press/Taylor & Francis; 2015. Chapter 25. PMID: 26269925 Free Books & Documents. Review.
References
-
- Anderberg C., Pietras K. (2009). On the origin of cancer-associated fibroblasts. Oxfordshire, United Kingdom: Taylor and Francis. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

