Open Reading Frame 4 Is Not Essential in the Replication and Infection of Genotype 1 Hepatitis E Virus
- PMID: 36992492
- PMCID: PMC10052008
- DOI: 10.3390/v15030784
Open Reading Frame 4 Is Not Essential in the Replication and Infection of Genotype 1 Hepatitis E Virus
Abstract
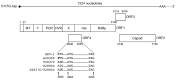
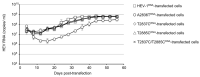
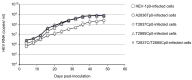
Genotype 1 hepatitis E virus (HEV-1), unlike other genotypes of HEV, has a unique small open reading frame known as ORF4 whose function is not yet known. ORF4 is located in an out-framed manner in the middle of ORF1, which encodes putative 90 to 158 amino acids depending on the strains. To explore the role of ORF4 in HEV-1 replication and infection, we cloned the complete genome of wild-type HEV-1 downstream of a T7 RNA polymerase promoter, and the following ORF4 mutant constructs were prepared: the first construct had TTG instead of the initiation codon ATG (A2836T), introducing an M→L mutation in ORF4 and a D→V mutation in ORF1. The second construct had ACG instead of the ATG codon (T2837C), introducing an M→T mutation in ORF4. The third construct had ACG instead of the second in-frame ATG codon (T2885C), introducing an M→T mutation in ORF4. The fourth construct contained two mutations (T2837C and T2885C) accompanying two M→T mutations in ORF4. For the latter three constructs, the accompanied mutations introduced in ORF1 were all synonymous changes. The capped entire genomic RNAs were generated by in vitro transcription and used to transfect PLC/PRF/5 cells. Three mRNAs containing synonymous mutations in ORF1, i.e., T2837CRNA, T2885CRNA, and T2837C/T2885CRNA, replicated normally in PLC/PRF/5 cells and generated infectious viruses that successfully infected Mongolian gerbils as the wild-type HEV-1 did. In contrast, the mutant RNA, i.e., A2836TRNA, accompanying an amino acid change (D937V) in ORF1 generated infectious viruses upon transfection, but they replicated slower than the wild-type HEV-1 and failed to infect Mongolian gerbils. No putative viral protein(s) derived from ORF4 were detected in the wild-type HEV-1- as well as the mutant virus-infected PLC/PRF/5 cells by Western blot analysis using a high-titer anti-HEV-1 IgG antibody. These results demonstrated that the ORF4-defective HEV-1s had the ability to replicate in the cultured cells, and that these defective viruses had the ability to infect Mongolian gerbils unless the overlapping ORF1 was accompanied by non-synonymous mutation(s), confirming that ORF4 is not essential in the replication and infection of HEV-1.
Keywords: HEV-1 mutant; Mongolian gerbil; ORF4 mutant; PLC/PRF/5 cells; genotype 1; hepatitis E virus; reverse genetic system.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Ectopic Expression of Genotype 1 Hepatitis E Virus ORF4 Increases Genotype 3 HEV Viral Replication in Cell Culture.Viruses. 2021 Jan 7;13(1):75. doi: 10.3390/v13010075. Viruses. 2021. PMID: 33430442 Free PMC article.
-
An analysis of two open reading frames (ORF3 and ORF4) of rat hepatitis E virus genome using its infectious cDNA clones with mutations in ORF3 or ORF4.Virus Res. 2018 Apr 2;249:16-30. doi: 10.1016/j.virusres.2018.02.014. Epub 2018 Feb 19. Virus Res. 2018. PMID: 29471051
-
Analysis of codon usage patterns in open reading frame 4 of hepatitis E viruses.Beni Suef Univ J Basic Appl Sci. 2022;11(1):65. doi: 10.1186/s43088-022-00244-w. Epub 2022 May 10. Beni Suef Univ J Basic Appl Sci. 2022. PMID: 35573872 Free PMC article.
-
Hepatitis E Virus Replication.Viruses. 2019 Aug 6;11(8):719. doi: 10.3390/v11080719. Viruses. 2019. PMID: 31390784 Free PMC article. Review.
-
[Cell culture system for hepatitis E virus].Uirusu. 2010 Jun;60(1):93-104. doi: 10.2222/jsv.60.93. Uirusu. 2010. PMID: 20848869 Review. Japanese.
Cited by
-
Rocahepevirus ratti as an Emerging Cause of Acute Hepatitis Worldwide.Microorganisms. 2023 Dec 16;11(12):2996. doi: 10.3390/microorganisms11122996. Microorganisms. 2023. PMID: 38138140 Free PMC article. Review.
-
Strain- and Subtype-Specific Replication of Genotype 3 Hepatitis E Viruses in Mongolian Gerbils.Viruses. 2024 Oct 12;16(10):1605. doi: 10.3390/v16101605. Viruses. 2024. PMID: 39459937 Free PMC article.
References
-
- Meng X.J., Anderson D.A., Arankalle V.A., Emerson S.U., Harrison T.J., Jameel S., Okamoto H. Hepeviridae. In: King A.M.A., Michael J., Carstens E.B., Lefkowitz E.J., editors. Virus Taxonomy: Ninth Report of the ICTV. Elsevier; London, UK: Academic Press; London, UK: 2012. pp. 1021–1028.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

