Gold Nanoparticle-Based Plasmonic Biosensors
- PMID: 36979623
- PMCID: PMC10046074
- DOI: 10.3390/bios13030411
Gold Nanoparticle-Based Plasmonic Biosensors
Abstract
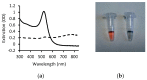
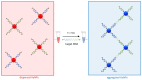
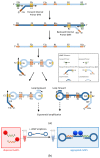
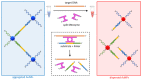
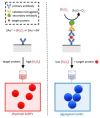
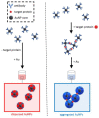
One of the emerging technologies in molecular diagnostics of the last two decades is the use of gold nanoparticles (AuNPs) for biosensors. AuNPs can be functionalized with various biomolecules, such as nucleic acids or antibodies, to recognize and bind to specific targets. AuNPs present unique optical properties, such as their distinctive plasmonic band, which confers a bright-red color to AuNP solutions, and their extremely high extinction coefficient, which makes AuNPs detectable by the naked eye even at low concentrations. Ingenious molecular mechanisms triggered by the presence of a target analyte can change the colloidal status of AuNPs from dispersed to aggregated, with a subsequent visible change in color of the solution due to the loss of the characteristic plasmonic band. This review describes how the optical properties of AuNPs have been exploited for the design of plasmonic biosensors that only require the simple mixing of reagents combined with a visual readout and focuses on the molecular mechanisms involved. This review illustrates selected examples of AuNP-based plasmonic biosensors and promising approaches for the point-of-care testing of various analytes, spanning from the viral RNA of SARS-CoV-2 to the molecules that give distinctive flavor and color to aged whisky.
Keywords: molecular diagnostics; naked-eye detection; point-of-care testing.
Conflict of interest statement
The author declares no conflict of interest.
Figures


Similar articles
-
Plasmon color-preserved gold nanoparticle clusters for high sensitivity detection of SARS-CoV-2 based on lateral flow immunoassay.Biosens Bioelectron. 2022 Jun 1;205:114094. doi: 10.1016/j.bios.2022.114094. Epub 2022 Feb 17. Biosens Bioelectron. 2022. PMID: 35202985 Free PMC article.
-
Colorimetric Detection of SARS-CoV-2 Using Plasmonic Biosensors and Smartphones.ACS Appl Mater Interfaces. 2022 Dec 14;14(49):54527-54538. doi: 10.1021/acsami.2c15407. Epub 2022 Dec 1. ACS Appl Mater Interfaces. 2022. PMID: 36454041
-
Colorimetric Test for Fast Detection of SARS-CoV-2 in Nasal and Throat Swabs.ACS Sens. 2020 Oct 23;5(10):3043-3048. doi: 10.1021/acssensors.0c01742. Epub 2020 Oct 1. ACS Sens. 2020. PMID: 32989986 Free PMC article.
-
A Review on the Development of Gold and Silver Nanoparticles-Based Biosensor as a Detection Strategy of Emerging and Pathogenic RNA Virus.Sensors (Basel). 2021 Jul 28;21(15):5114. doi: 10.3390/s21155114. Sensors (Basel). 2021. PMID: 34372350 Free PMC article. Review.
-
Recent advances in gold nanoparticles-based biosensors for food safety detection.Biosens Bioelectron. 2021 May 1;179:113076. doi: 10.1016/j.bios.2021.113076. Epub 2021 Feb 7. Biosens Bioelectron. 2021. PMID: 33601132 Review.
Cited by
-
Ultrasmall ATP-Coated Gold Nanoparticles Specifically Bind to Non-Hybridized Regions in DNA.Nanomaterials (Basel). 2023 Dec 5;13(24):3080. doi: 10.3390/nano13243080. Nanomaterials (Basel). 2023. PMID: 38132978 Free PMC article.
-
Research Progress of Polysaccharide-Gold Nanocomplexes in Drug Delivery.Pharmaceutics. 2024 Jan 9;16(1):88. doi: 10.3390/pharmaceutics16010088. Pharmaceutics. 2024. PMID: 38258099 Free PMC article. Review.
-
Using AuNPs-DNA Walker with Fluorophores Detects the Hepatitis Virus Rapidly.Biosensors (Basel). 2024 Jul 29;14(8):370. doi: 10.3390/bios14080370. Biosensors (Basel). 2024. PMID: 39194599 Free PMC article.
-
Biotechnological advances in microbial synthesis of gold nanoparticles: Optimizations and applications.3 Biotech. 2024 Nov;14(11):263. doi: 10.1007/s13205-024-04110-7. Epub 2024 Oct 7. 3 Biotech. 2024. PMID: 39387004 Review.
-
Sustainable and Reusable Modified Membrane Based on Green Gold Nanoparticles for Efficient Methylene Blue Water Decontamination by a Photocatalytic Process.Nanomaterials (Basel). 2024 Oct 8;14(19):1611. doi: 10.3390/nano14191611. Nanomaterials (Basel). 2024. PMID: 39404338 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

