Laminin switches terminal differentiation fate of human trophoblast stem cells under chemically defined culture conditions
- PMID: 36972789
- PMCID: PMC10176266
- DOI: 10.1016/j.jbc.2023.104650
Laminin switches terminal differentiation fate of human trophoblast stem cells under chemically defined culture conditions
Abstract
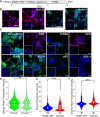
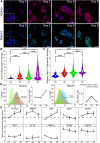
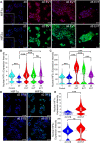
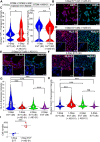
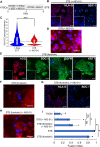
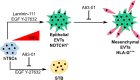
Human trophoblast stem cells (hTSCs) have emerged as a powerful tool to model early placental development in vitro. Analogous to the epithelial cytotrophoblast in the placenta, hTSCs can differentiate into cells of the extravillous trophoblast (EVT) lineage or the multinucleate syncytiotrophoblast (STB). Here we present a chemically defined culture system for STB and EVT differentiation of hTSCs. Notably, in contrast to current approaches, we neither utilize forskolin for STB formation nor transforming growth factor-beta (TGFβ) inhibitors or a passage step for EVT differentiation. Strikingly, the presence of a single additional extracellular cue-laminin-111-switched the terminal differentiation of hTSCs from STB to the EVT lineage under these conditions. In the absence of laminin-111, STB formation occurred, with cell fusion comparable to that obtained with differentiation mediated by forskolin; however, in the presence of laminin-111, hTSCs differentiated to the EVT lineage. Protein expression of nuclear hypoxia-inducible factors (HIF1α and HIF2α) was upregulated during EVT differentiation mediated by laminin-111 exposure. A heterogeneous mixture of Notch1+ EVTs in colonies and HLA-G+ single-cell EVTs were obtained without a passage step, reminiscent of heterogeneity in vivo. Further analysis showed that inhibition of TGFβ signaling affected both STB and EVT differentiation mediated by laminin-111 exposure. TGFβ inhibition during EVT differentiation resulted in decreased HLA-G expression and increased Notch1 expression. On the other hand, TGFβ inhibition prevented STB formation. The chemically defined culture system for hTSC differentiation established herein facilitates quantitative analysis of heterogeneity that arises during hTSC differentiation and will enable mechanistic studies in vitro.
Keywords: differentiation; extravillous trophoblast; placenta; syncytiotrophoblast; trophoblast; trophoblast stem cells.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures


Similar articles
-
Generation of Human Trophoblast Stem Cell-Dependent Placental In Vitro Models.Methods Mol Biol. 2024;2767:43-52. doi: 10.1007/7651_2022_463. Methods Mol Biol. 2024. PMID: 36515896
-
Derivation of trophoblast stem cells from naïve human pluripotent stem cells.Elife. 2020 Feb 12;9:e52504. doi: 10.7554/eLife.52504. Elife. 2020. PMID: 32048992 Free PMC article.
-
Human pluripotent stem cells as a model of trophoblast differentiation in both normal development and disease.Proc Natl Acad Sci U S A. 2016 Jul 5;113(27):E3882-91. doi: 10.1073/pnas.1604747113. Epub 2016 Jun 20. Proc Natl Acad Sci U S A. 2016. PMID: 27325764 Free PMC article.
-
Trophoblast lineage specification, differentiation and their regulation by oxygen tension.J Endocrinol. 2018 Jan;236(1):R43-R56. doi: 10.1530/JOE-17-0402. J Endocrinol. 2018. PMID: 29259074 Free PMC article. Review.
-
WNT and NOTCH signaling in human trophoblast development and differentiation.Cell Mol Life Sci. 2022 May 13;79(6):292. doi: 10.1007/s00018-022-04285-3. Cell Mol Life Sci. 2022. PMID: 35562545 Free PMC article. Review.
References
-
- Aplin J.D. Developmental cell biology of human villous trophoblast: current research problems. Int. J. Dev. Biol. 2010;54:323–329. - PubMed
-
- Knöfler M., Vasicek R., Schreiber M. Key regulatory transcription factors involved in placental trophoblast development - a review. Placenta. 2001;22:S83–S92. - PubMed
-
- Loregger T., Pollheimer J., Knöfler M. Regulatory transcription factors controlling function and differentiation of human trophoblast - a review. Placenta. 2003;24:S104–S110. - PubMed
-
- Caniggia I., Winter J., Lye S.J., Post M. Oxygen and placental development during the first trimester: implications for the pathophysiology of pre-eclampsia. Placenta. 2000;21:S25–S30. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

