Single nucleus multiomics identifies ZEB1 and MAFB as candidate regulators of Alzheimer's disease-specific cis-regulatory elements
- PMID: 36950385
- PMCID: PMC10025452
- DOI: 10.1016/j.xgen.2023.100263
Single nucleus multiomics identifies ZEB1 and MAFB as candidate regulators of Alzheimer's disease-specific cis-regulatory elements
Abstract
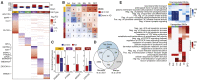
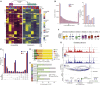
Cell type-specific transcriptional differences between brain tissues from donors with Alzheimer's disease (AD) and unaffected controls have been well documented, but few studies have rigorously interrogated the regulatory mechanisms responsible for these alterations. We performed single nucleus multiomics (snRNA-seq plus snATAC-seq) on 105,332 nuclei isolated from cortical tissues from 7 AD and 8 unaffected donors to identify candidate cis-regulatory elements (CREs) involved in AD-associated transcriptional changes. We detected 319,861 significant correlations, or links, between gene expression and cell type-specific transposase accessible regions enriched for active CREs. Among these, 40,831 were unique to AD tissues. Validation experiments confirmed the activity of many regions, including several candidate regulators of APP expression. We identified ZEB1 and MAFB as candidate transcription factors playing important roles in AD-specific gene regulation in neurons and microglia, respectively. Microglia links were globally enriched for heritability of AD risk and previously identified active regulatory regions.
Keywords: APP; Alzheimer’s; MAFB; ZEB1; cis-regulatory element; microglia; multiomics; neuron; single cell; transcription factor.
© 2023 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Single-nucleus multiome analysis of human cerebellum in Alzheimer's disease-related dementia.Res Sq [Preprint]. 2024 Aug 16:rs.3.rs-4871032. doi: 10.21203/rs.3.rs-4871032/v1. Res Sq. 2024. PMID: 39184089 Free PMC article. Preprint.
-
Integrative single-nucleus multi-omics analysis prioritizes candidate cis and trans regulatory networks and their target genes in Alzheimer's disease brains.Cell Biosci. 2023 Oct 3;13(1):185. doi: 10.1186/s13578-023-01120-5. Cell Biosci. 2023. PMID: 37789374 Free PMC article.
-
Single-nucleus multiomics reveals the disrupted regulatory programs in three brain regions of sporadic early-onset Alzheimer's disease.bioRxiv [Preprint]. 2024 Jun 29:2024.06.25.600720. doi: 10.1101/2024.06.25.600720. bioRxiv. 2024. PMID: 38979371 Free PMC article. Preprint.
-
Transcriptional Networks of Microglia in Alzheimer's Disease and Insights into Pathogenesis.Genes (Basel). 2019 Oct 12;10(10):798. doi: 10.3390/genes10100798. Genes (Basel). 2019. PMID: 31614849 Free PMC article. Review.
-
Impact of non-neuronal cells in Alzheimer's disease from a single-nucleus profiling perspective.Front Cell Neurosci. 2023 Jun 14;17:1208122. doi: 10.3389/fncel.2023.1208122. eCollection 2023. Front Cell Neurosci. 2023. PMID: 37388411 Free PMC article. Review.
Cited by
-
Neuronal MAPT expression is mediated by long-range interactions with cis-regulatory elements.Am J Hum Genet. 2024 Feb 1;111(2):259-279. doi: 10.1016/j.ajhg.2023.12.015. Epub 2024 Jan 16. Am J Hum Genet. 2024. PMID: 38232730 Free PMC article.
-
The broken Alzheimer's disease genome.Cell Genom. 2024 May 8;4(5):100555. doi: 10.1016/j.xgen.2024.100555. Epub 2024 May 1. Cell Genom. 2024. PMID: 38697121 Free PMC article. Review.
-
Cell-type-specific mapping of enhancers and target genes from single-cell multimodal data.bioRxiv [Preprint]. 2024 Sep 26:2024.09.24.614814. doi: 10.1101/2024.09.24.614814. bioRxiv. 2024. PMID: 39386519 Free PMC article. Preprint.
-
Integration of human organoids single-cell transcriptomic profiles and human genetics repurposes critical cell type-specific drug targets for severe COVID-19.Cell Prolif. 2024 Mar;57(3):e13558. doi: 10.1111/cpr.13558. Epub 2023 Oct 8. Cell Prolif. 2024. PMID: 37807299 Free PMC article.
-
Neuronal enhancers fine-tune adaptive circuit plasticity.Neuron. 2024 Sep 25;112(18):3043-3057. doi: 10.1016/j.neuron.2024.08.002. Epub 2024 Aug 28. Neuron. 2024. PMID: 39208805 Review.
References
-
- Kunkle B.W., Grenier-Boley B., Sims R., Bis J.C., Damotte V., Naj A.C., Boland A., Vronskaya M., van der Lee S.J., Amlie-Wolf A., et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat. Genet. 2019;51:414–430. - PMC - PubMed
-
- Schwartzentruber J., Cooper S., Liu J.Z., Barrio-Hernandez I., Bello E., Kumasaka N., Young A.M.H., Franklin R.J.M., Johnson T., Estrada K., et al. Genome-wide meta-analysis, fine-mapping and integrative prioritization implicate new Alzheimer’s disease risk genes. Nat. Genet. 2021;53:392–402. - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

