A swapped genetic code prevents viral infections and gene transfer
- PMID: 36922599
- PMCID: PMC10151025
- DOI: 10.1038/s41586-023-05824-z
A swapped genetic code prevents viral infections and gene transfer
Abstract
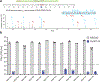
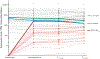
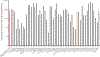
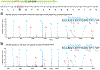
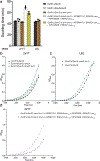
Engineering the genetic code of an organism has been proposed to provide a firewall from natural ecosystems by preventing viral infections and gene transfer1-6. However, numerous viruses and mobile genetic elements encode parts of the translational apparatus7-9, potentially rendering a genetic-code-based firewall ineffective. Here we show that such mobile transfer RNAs (tRNAs) enable gene transfer and allow viral replication in Escherichia coli despite the genome-wide removal of 3 of the 64 codons and the previously essential cognate tRNA and release factor genes. We then establish a genetic firewall by discovering viral tRNAs that provide exceptionally efficient codon reassignment allowing us to develop cells bearing an amino acid-swapped genetic code that reassigns two of the six serine codons to leucine during translation. This amino acid-swapped genetic code renders cells resistant to viral infections by mistranslating viral proteomes and prevents the escape of synthetic genetic information by engineered reliance on serine codons to produce leucine-requiring proteins. As these cells may have a selective advantage over wild organisms due to virus resistance, we also repurpose a third codon to biocontain this virus-resistant host through dependence on an amino acid not found in nature10. Our results may provide the basis for a general strategy to make any organism safely resistant to all natural viruses and prevent genetic information flow into and out of genetically modified organisms.
© 2023. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing Interests statement
The authors declare competing financial interests. Harvard Medical School has filed a provisional patent application related to this work on which A.N., S.V., and G.M.C. are listed as inventors. M.L., K.C., and F.H. are employed by GenScript USA Inc., but the company had no role in designing or executing experiments. G.M.C. is a founder of the following companies in which he has related financial interests: GRO Biosciences, EnEvolv (Ginkgo Bioworks), and 64x Bio. Other potentially relevant financial interests of G.M.C. are listed at
Figures

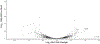
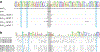

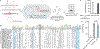



Comment in
-
Synthetic bacterial genome upgraded for viral defence and biocontainment.Nature. 2023 Mar;615(7953):592-594. doi: 10.1038/d41586-023-00702-0. Nature. 2023. PMID: 36922657 No abstract available.
-
Retrofitted E. coli genome confers phage resistance.Nat Biotechnol. 2023 Apr;41(4):470. doi: 10.1038/s41587-023-01764-1. Nat Biotechnol. 2023. PMID: 37069393 No abstract available.
Similar articles
-
An Amino Acid-Swapped Genetic Code.ACS Synth Biol. 2020 Oct 16;9(10):2703-2713. doi: 10.1021/acssynbio.0c00196. Epub 2020 Sep 18. ACS Synth Biol. 2020. PMID: 32882137
-
Recoded organisms engineered to depend on synthetic amino acids.Nature. 2015 Feb 5;518(7537):89-93. doi: 10.1038/nature14095. Epub 2015 Jan 21. Nature. 2015. PMID: 25607356 Free PMC article.
-
Expansion of the genetic code through reassignment of redundant sense codons using fully modified tRNA.Nucleic Acids Res. 2022 Oct 28;50(19):11374-11386. doi: 10.1093/nar/gkac846. Nucleic Acids Res. 2022. PMID: 36300637 Free PMC article.
-
Recent advances in genetic code engineering in Escherichia coli.Curr Opin Biotechnol. 2012 Oct;23(5):751-7. doi: 10.1016/j.copbio.2011.12.027. Epub 2012 Jan 9. Curr Opin Biotechnol. 2012. PMID: 22237016 Review.
-
Engineering the Genetic Code in Cells and Animals: Biological Considerations and Impacts.Acc Chem Res. 2017 Nov 21;50(11):2767-2775. doi: 10.1021/acs.accounts.7b00376. Epub 2017 Oct 6. Acc Chem Res. 2017. PMID: 28984438 Free PMC article. Review.
Cited by
-
A robust yeast biocontainment system with two-layered regulation switch dependent on unnatural amino acid.Nat Commun. 2023 Oct 14;14(1):6487. doi: 10.1038/s41467-023-42358-4. Nat Commun. 2023. PMID: 37838746 Free PMC article.
-
Ser/Leu-swapped cell-free translation system constructed with natural/in vitro transcribed-hybrid tRNA set.Nat Commun. 2024 May 16;15(1):4143. doi: 10.1038/s41467-024-48056-z. Nat Commun. 2024. PMID: 38755134 Free PMC article.
-
Synthetic bacterial genome upgraded for viral defence and biocontainment.Nature. 2023 Mar;615(7953):592-594. doi: 10.1038/d41586-023-00702-0. Nature. 2023. PMID: 36922657 No abstract available.
-
Identification of HDV-like theta ribozymes involved in tRNA-based recoding of gut bacteriophages.Nat Commun. 2024 Feb 20;15(1):1559. doi: 10.1038/s41467-024-45653-w. Nat Commun. 2024. PMID: 38378708 Free PMC article.
-
Engineering bacterial theranostics: from logic gates to in vivo applications.Front Bioeng Biotechnol. 2024 Sep 18;12:1437301. doi: 10.3389/fbioe.2024.1437301. eCollection 2024. Front Bioeng Biotechnol. 2024. PMID: 39359265 Free PMC article. Review.
References
-
- Church GM & Regis E Regenesis: How Synthetic Biology Will Reinvent Nature and Ourselves. (Basic Books, 2014).
-
- Ostrov N et al. Design, synthesis, and testing toward a 57-codon genome. Science 353, 819–822 (2016). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

