Structural basis for the Rad6 activation by the Bre1 N-terminal domain
- PMID: 36912886
- PMCID: PMC10036116
- DOI: 10.7554/eLife.84157
Structural basis for the Rad6 activation by the Bre1 N-terminal domain
Abstract
The mono-ubiquitination of the histone protein H2B (H2Bub1) is a highly conserved histone post-translational modification that plays critical roles in many fundamental processes. In yeast, this modification is catalyzed by the conserved Bre1-Rad6 complex. Bre1 contains a unique N-terminal Rad6-binding domain (RBD), how it interacts with Rad6 and contributes to the H2Bub1 catalysis is unclear. Here, we present crystal structure of the Bre1 RBD-Rad6 complex and structure-guided functional studies. Our structure provides a detailed picture of the interaction between the dimeric Bre1 RBD and a single Rad6 molecule. We further found that the interaction stimulates Rad6's enzymatic activity by allosterically increasing its active site accessibility and likely contribute to the H2Bub1 catalysis through additional mechanisms. In line with these important functions, we found that the interaction is crucial for multiple H2Bub1-regulated processes. Our study provides molecular insights into the H2Bub1 catalysis.
Keywords: Bre1; Rad6; histone; molecular biophysics; post-translational modification; structural biology; ubiquitin.
© 2023, Shi, Zhao, Zhang et al.
Conflict of interest statement
MS, JZ, SZ, WH, ML, XB, WZ, KZ, XC, SX No competing interests declared
Figures



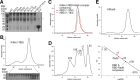
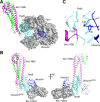








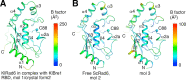
Update of
- doi: 10.1101/2022.10.23.513400
Similar articles
-
The Bre1/Rad6 machinery: writing the central histone ubiquitin mark on H2B and beyond.Chromosome Res. 2020 Dec;28(3-4):247-258. doi: 10.1007/s10577-020-09640-3. Epub 2020 Sep 7. Chromosome Res. 2020. PMID: 32895784 Review.
-
Structural basis for the role of C-terminus acidic tail of Saccharomyces cerevisiae ubiquitin-conjugating enzyme (Rad6) in E3 ligase (Bre1) mediated recognition of histones.Int J Biol Macromol. 2024 Jan;254(Pt 2):127717. doi: 10.1016/j.ijbiomac.2023.127717. Epub 2023 Nov 2. Int J Biol Macromol. 2024. PMID: 37923031
-
Rad6-Bre1-mediated H2B ubiquitination regulates telomere replication by promoting telomere-end resection.Nucleic Acids Res. 2017 Apr 7;45(6):3308-3322. doi: 10.1093/nar/gkx101. Nucleic Acids Res. 2017. PMID: 28180293 Free PMC article.
-
Structure and functional determinants of Rad6-Bre1 subunits in the histone H2B ubiquitin-conjugating complex.Nucleic Acids Res. 2023 Mar 21;51(5):2117-2136. doi: 10.1093/nar/gkad012. Nucleic Acids Res. 2023. PMID: 36715322 Free PMC article.
-
The role of RAD6 in recombinational repair, checkpoints and meiosis via histone modification.DNA Repair (Amst). 2009 Apr 5;8(4):470-82. doi: 10.1016/j.dnarep.2009.01.007. Epub 2009 Feb 18. DNA Repair (Amst). 2009. PMID: 19230796 Review.
Cited by
-
Histone H2B ubiquitylation: Connections to transcription and effects on chromatin structure.Biochim Biophys Acta Gene Regul Mech. 2024 Jun;1867(2):195018. doi: 10.1016/j.bbagrm.2024.195018. Epub 2024 Feb 6. Biochim Biophys Acta Gene Regul Mech. 2024. PMID: 38331024 Review.
-
Paf1 complex subunit Rtf1 stimulates H2B ubiquitylation by interacting with the highly conserved N-terminal helix of Rad6.Proc Natl Acad Sci U S A. 2023 May 30;120(22):e2220041120. doi: 10.1073/pnas.2220041120. Epub 2023 May 22. Proc Natl Acad Sci U S A. 2023. PMID: 37216505 Free PMC article.
References
-
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive python-based system for macromolecular structure solution. Acta Crystallographica. Section D, Biological Crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

