HuR modulation counteracts lipopolysaccharide response in murine macrophages
- PMID: 36912171
- PMCID: PMC10110401
- DOI: 10.1242/dmm.050120
HuR modulation counteracts lipopolysaccharide response in murine macrophages
Abstract
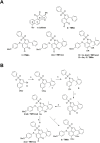
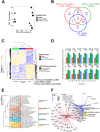
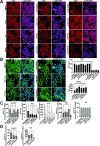
Lipopolysaccharide (LPS) exposure to macrophages induces an inflammatory response, which is regulated at the transcriptional and post-transcriptional levels. HuR (ELAVL1) is an RNA-binding protein that regulates cytokines and chemokines transcripts containing AU/U-rich elements (AREs) and mediates the LPS-induced response. Here, we show that small-molecule tanshinone mimics (TMs) inhibiting HuR-RNA interaction counteract LPS stimulus in macrophages. TMs exist in solution in keto-enolic tautomerism, and molecular dynamic calculations showed the ortho-quinone form inhibiting binding of HuR to mRNA targets. TM activity was lost in vitro by blocking the diphenolic reduced form as a diacetate, but resulted in prodrug-like activity in vivo. RNA and ribonucleoprotein immunoprecipitation sequencing revealed that LPS induces a strong coupling between differentially expressed genes and HuR-bound genes, and TMs reduced such interactions. TMs decreased the association of HuR with genes involved in chemotaxis and immune response, including Cxcl10, Il1b and Cd40, reducing their expression and protein secretion in primary murine bone marrow-derived macrophages and in an LPS-induced peritonitis model. Overall, TMs show anti-inflammatory properties in vivo and suggest HuR as a potential therapeutic target for inflammation-related diseases.
Keywords: Anti-inflammatory agents; ELAVL1; HuR; LPS; RIP-seq; Tanshinone mimics.
© 2023. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures




Similar articles
-
Novel, soluble 3-heteroaryl-substituted tanshinone mimics attenuate the inflammatory response in murine macrophages.Sci Rep. 2024 Oct 18;14(1):24501. doi: 10.1038/s41598-024-73309-8. Sci Rep. 2024. PMID: 39424621 Free PMC article.
-
Chronic ethanol exposure increases the binding of HuR to the TNFalpha 3'-untranslated region in macrophages.J Biol Chem. 2003 Oct 3;278(40):38333-41. doi: 10.1074/jbc.M304566200. Epub 2003 Jul 21. J Biol Chem. 2003. PMID: 12876290 Free PMC article.
-
Induction of Glucocorticoid-induced Leucine Zipper (GILZ) Contributes to Anti-inflammatory Effects of the Natural Product Curcumin in Macrophages.J Biol Chem. 2016 Oct 28;291(44):22949-22960. doi: 10.1074/jbc.M116.733253. Epub 2016 Sep 14. J Biol Chem. 2016. PMID: 27629417 Free PMC article.
-
Multiple functions of the RNA-binding protein HuR in cancer progression, treatment responses and prognosis.Int J Mol Sci. 2013 May 10;14(5):10015-41. doi: 10.3390/ijms140510015. Int J Mol Sci. 2013. PMID: 23665903 Free PMC article. Review.
-
Dysregulation of TTP and HuR plays an important role in cancers.Tumour Biol. 2016 Nov;37(11):14451-14461. doi: 10.1007/s13277-016-5397-z. Epub 2016 Sep 19. Tumour Biol. 2016. PMID: 27644249 Review.
Cited by
-
Multiple roles for AU-rich RNA binding proteins in the development of haematologic malignancies and their resistance to chemotherapy.RNA Biol. 2024 Jan;21(1):1-17. doi: 10.1080/15476286.2024.2346688. Epub 2024 May 27. RNA Biol. 2024. PMID: 38798162 Free PMC article. Review.
-
Phosphorylation at the disordered N-end makes HuR accumulate and dimerize in the cytoplasm.Nucleic Acids Res. 2024 Aug 12;52(14):8552-8565. doi: 10.1093/nar/gkae564. Nucleic Acids Res. 2024. PMID: 38966993 Free PMC article.
-
Novel, soluble 3-heteroaryl-substituted tanshinone mimics attenuate the inflammatory response in murine macrophages.Sci Rep. 2024 Oct 18;14(1):24501. doi: 10.1038/s41598-024-73309-8. Sci Rep. 2024. PMID: 39424621 Free PMC article.
References
-
- Abraham, M. J., Murtola, T., Schulz, R., Páll, S., Smith, J. C., Hess, B. and Lindahl, E. (2015). GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1-2, 19-25. 10.1016/j.softx.2015.06.001 - DOI
-
- Assoni, G., La Pietra, V., Digilio, R., Ciani, C., Licata, N. V., Micaelli, M., Facen, E., Tomaszewska, W., Cerofolini, L., Pérez-Ràfols, A.et al. (2022). HuR-targeted agents: an insight into medicinal chemistry, biophysical, computational studies and pharmacological effects on cancer models. Adv. Drug Deliv. Rev. 181, 114088. 10.1016/J.ADDR.2021.114088 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

