The ubiquitin-specific protease USP36 SUMOylates EXOSC10 and promotes the nucleolar RNA exosome function in rRNA processing
- PMID: 36912080
- PMCID: PMC10164564
- DOI: 10.1093/nar/gkad140
The ubiquitin-specific protease USP36 SUMOylates EXOSC10 and promotes the nucleolar RNA exosome function in rRNA processing
Abstract
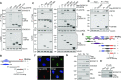
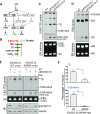
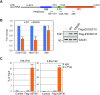
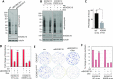
The RNA exosome is an essential 3' to 5' exoribonuclease complex that mediates degradation, processing and quality control of virtually all eukaryotic RNAs. The nucleolar RNA exosome, consisting of a nine-subunit core and a distributive 3' to 5' exonuclease EXOSC10, plays a critical role in processing and degrading nucleolar RNAs, including pre-rRNA. However, how the RNA exosome is regulated in the nucleolus is poorly understood. Here, we report that the nucleolar ubiquitin-specific protease USP36 is a novel regulator of the nucleolar RNA exosome. USP36 binds to the RNA exosome through direct interaction with EXOSC10 in the nucleolus. Interestingly, USP36 does not significantly regulate the levels of EXOSC10 and other tested exosome subunits. Instead, it mediates EXOSC10 SUMOylation at lysine (K) 583. Mutating K583 impaired the binding of EXOSC10 to pre-rRNAs, and the K583R mutant failed to rescue the defects in rRNA processing and cell growth inhibition caused by knockdown of endogenous EXOSC10. Furthermore, EXOSC10 SUMOylation is markedly reduced in cells in response to perturbation of ribosomal biogenesis. Together, these results suggest that USP36 acts as a SUMO ligase to promote EXOSC10 SUMOylation critical for the RNA exosome function in ribosome biogenesis.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



Similar articles
-
Hypoxia-driven deSUMOylation of EXOSC10 promotes adaptive changes in the transcriptome profile.Cell Mol Life Sci. 2024 Jan 27;81(1):58. doi: 10.1007/s00018-023-05035-9. Cell Mol Life Sci. 2024. PMID: 38279024 Free PMC article.
-
Cooling-induced SUMOylation of EXOSC10 down-regulates ribosome biogenesis.RNA. 2016 Apr;22(4):623-35. doi: 10.1261/rna.054411.115. Epub 2016 Feb 8. RNA. 2016. PMID: 26857222 Free PMC article.
-
EXOSC10/Rrp6 is essential for the eight-cell embryo/morula transition.Dev Biol. 2022 Mar;483:58-65. doi: 10.1016/j.ydbio.2021.12.010. Epub 2021 Dec 26. Dev Biol. 2022. PMID: 34965385
-
Regulation of the conserved 3'-5' exoribonuclease EXOSC10/Rrp6 during cell division, development and cancer.Biol Rev Camb Philos Soc. 2021 Aug;96(4):1092-1113. doi: 10.1111/brv.12693. Epub 2021 Feb 18. Biol Rev Camb Philos Soc. 2021. PMID: 33599082 Review.
-
The eukaryotic RNA exosome.Curr Opin Struct Biol. 2014 Feb;24:132-40. doi: 10.1016/j.sbi.2014.01.011. Epub 2014 Feb 11. Curr Opin Struct Biol. 2014. PMID: 24525139 Free PMC article. Review.
Cited by
-
USP36 SUMOylates Las1L and Promotes Its Function in Pre-Ribosomal RNA ITS2 Processing.Cancer Res Commun. 2024 Oct 1;4(10):2835-2845. doi: 10.1158/2767-9764.CRC-24-0312. Cancer Res Commun. 2024. PMID: 39356143 Free PMC article.
-
The Emerging Role of Ubiquitin-Specific Protease 36 (USP36) in Cancer and Beyond.Biomolecules. 2024 May 12;14(5):572. doi: 10.3390/biom14050572. Biomolecules. 2024. PMID: 38785979 Free PMC article. Review.
-
SUMOylation regulation of ribosome biogenesis: Emerging roles for USP36.Front RNA Res. 2024;2:1389104. doi: 10.3389/frnar.2024.1389104. Epub 2024 Apr 3. Front RNA Res. 2024. PMID: 38764604 Free PMC article.
-
In the moonlight: non-catalytic functions of ubiquitin and ubiquitin-like proteases.Front Mol Biosci. 2024 Feb 22;11:1349509. doi: 10.3389/fmolb.2024.1349509. eCollection 2024. Front Mol Biosci. 2024. PMID: 38455765 Free PMC article. Review.
-
Hypoxia-driven deSUMOylation of EXOSC10 promotes adaptive changes in the transcriptome profile.Cell Mol Life Sci. 2024 Jan 27;81(1):58. doi: 10.1007/s00018-023-05035-9. Cell Mol Life Sci. 2024. PMID: 38279024 Free PMC article.
References
-
- Kressler D., Hurt E., Bassler J.. A puzzle of life: crafting ribosomal subunits. Trends Biochem. Sci. 2017; 42:640–654. - PubMed
-
- Rodnina M.V., Wintermeyer W.. Recent mechanistic insights into eukaryotic ribosomes. Curr. Opin. Cell Biol. 2009; 21:435–443. - PubMed
-
- Kilchert C., Wittmann S., Vasiljeva L.. The regulation and functions of the nuclear RNA exosome complex. Nat. Rev. Mol. Cell Biol. 2016; 17:227–239. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

