Acral Melanoma Is Infiltrated with cDC1s and Functional Exhausted CD8 T Cells Similar to the Cutaneous Melanoma of Sun-Exposed Skin
- PMID: 36902214
- PMCID: PMC10003718
- DOI: 10.3390/ijms24054786
Acral Melanoma Is Infiltrated with cDC1s and Functional Exhausted CD8 T Cells Similar to the Cutaneous Melanoma of Sun-Exposed Skin
Abstract
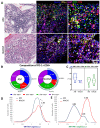
Acral melanoma (AM) is the most common melanoma in non-Caucasian populations, yet it remains largely understudied. As AM lacks the UV-radiation mutational signatures that characterize other cutaneous melanomas, it is considered devoid of immunogenicity and is rarely included in clinical trials assessing novel immunotherapeutic regimes aiming to recover the antitumor function of immune cells. We studied a Mexican cohort of melanoma patients from the Mexican Institute of Social Security (IMSS) (n = 38) and found an overrepresentation of AM (73.9%). We developed a multiparametric immunofluorescence technique coupled with a machine learning image analysis to evaluate the presence of conventional type 1 dendritic cells (cDC1) and CD8 T cells in the stroma of melanoma, two of the most relevant immune cell types for antitumor responses. We observed that both cell types infiltrate AM at similar and even higher levels than other cutaneous melanomas. Both melanoma types harbored programmed cell death protein 1 (PD-1+) CD8 T cells and PD-1 ligand (PD-L1+) cDC1s. Despite this, CD8 T cells appeared to preserve their effector function and expanding capacity as they expressed interferon-γ (IFN-γ) and KI-67. The density of cDC1s and CD8 T cells significantly decreased in advanced stage III and IV melanomas, supporting these cells' capacity to control tumor progression. These data also argue that AM could respond to anti-PD-1-PD-L1 immunotherapy.
Keywords: CD8; PD-1; PD-L1; acral; cDC1s; exhaustion; melanoma.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
TCF1-positive and TCF1-negative TRM CD8 T cell subsets and cDC1s orchestrate melanoma protection and immunotherapy response.J Immunother Cancer. 2024 Jul 5;12(7):e008739. doi: 10.1136/jitc-2023-008739. J Immunother Cancer. 2024. PMID: 38969523 Free PMC article.
-
In situ delivery of iPSC-derived dendritic cells with local radiotherapy generates systemic antitumor immunity and potentiates PD-L1 blockade in preclinical poorly immunogenic tumor models.J Immunother Cancer. 2021 May;9(5):e002432. doi: 10.1136/jitc-2021-002432. J Immunother Cancer. 2021. PMID: 34049930 Free PMC article.
-
Appraisal of clinicopathological prognosticators in advanced acral lentiginous melanoma with characterization of PD-L1 and CD8/CD4 immunoprofiles.Jpn J Clin Oncol. 2022 Sep 18;52(9):975-981. doi: 10.1093/jjco/hyac093. Jpn J Clin Oncol. 2022. PMID: 35662346
-
Tumoral PD-L1 expression in desmoplastic melanoma is associated with depth of invasion, tumor-infiltrating CD8 cytotoxic lymphocytes and the mixed cytomorphological variant.Mod Pathol. 2017 Mar;30(3):357-369. doi: 10.1038/modpathol.2016.210. Epub 2017 Jan 13. Mod Pathol. 2017. PMID: 28084337
-
Evolving cancer resistance to anti-PD-1/PD-L1 antibodies in melanoma: Comprehensive insights with future prospects.Crit Rev Oncol Hematol. 2024 Sep;201:104426. doi: 10.1016/j.critrevonc.2024.104426. Epub 2024 Jun 20. Crit Rev Oncol Hematol. 2024. PMID: 38908767 Review.
Cited by
-
TCF1-positive and TCF1-negative TRM CD8 T cell subsets and cDC1s orchestrate melanoma protection and immunotherapy response.J Immunother Cancer. 2024 Jul 5;12(7):e008739. doi: 10.1136/jitc-2023-008739. J Immunother Cancer. 2024. PMID: 38969523 Free PMC article.
References
-
- Lino-Silva L.S., Domínguez-Rodríguez J.A., Aguilar-Romero J.M., Martínez-Said H., Salcedo-Hernández R.A., García-Pérez L., Herrera-Gómez Á., Cuellar-Hubbe M. Melanoma in Mexico: Clinicopathologic Features in a Population with Predominance of Acral Lentiginous Subtype. Ann. Surg. Oncol. 2016;23:4189–4194. doi: 10.1245/s10434-016-5394-x. - DOI - PubMed
-
- Hernández J.F.G. Melanoma cutáneo (MC): Diagnóstico y tratamiento actuales. Gac. Med. Mex. 2014;150:175–182. - PubMed
-
- Barnes P.W., Robson T.M., Neale P.J., Williamson C.E., Zepp R.G., Madronich S., Wilson S.R., Andrady A.L., Heikkilä A.M., Bernhard G.H., et al. Environmental Effects of Stratospheric Ozone Depletion, UV Radiation, and Interactions with Climate Change: UNEP Environmental Effects Assessment Panel, Update 2021. Photochem. Photobiol. Sci. 2022;21:275–301. doi: 10.1007/s43630-022-00176-5. - DOI - PMC - PubMed
-
- Hodi F.S., Wolchok J.D., Schadendorf D., Larkin J., Long G.V., Qian X., Saci A., Young T.C., Srinivasan S., Chang H., et al. TMB and Inflammatory Gene Expression Associated with Clinical Outcomes Following Immunotherapy in Advanced Melanoma. Cancer Immunol. Res. 2021;9:1202–1213. doi: 10.1158/2326-6066.CIR-20-0983. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

