General Trends of the Camelidae Antibody VHHs Domain Dynamics
- PMID: 36901942
- PMCID: PMC10003728
- DOI: 10.3390/ijms24054511
General Trends of the Camelidae Antibody VHHs Domain Dynamics
Abstract
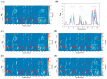
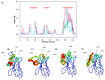
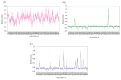
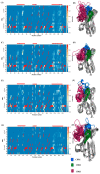
Conformational flexibility plays an essential role in antibodies' functional and structural stability. They facilitate and determine the strength of antigen-antibody interactions. Camelidae express an interesting subtype of single-chain antibody, named Heavy Chain only Antibody. They have only one N-terminal Variable domain (VHH) per chain, composed of Frameworks (FRs) and Complementarity Determining regions (CDRs) like their VH and VL counterparts in IgG. Even when expressed independently, VHH domains display excellent solubility and (thermo)stability, which helps them to retain their impressive interaction capabilities. Sequence and structural features of VHH domains contributing to these abilities have already been studied compared to classical antibodies. To have the broadest view and understand the changes in dynamics of these macromolecules, large-scale molecular dynamics simulations for a large number of non-redundant VHH structures have been performed for the first time. This analysis reveals the most prevalent movements in these domains. It reveals the four main classes of VHHs dynamics. Diverse local changes were observed in CDRs with various intensities. Similarly, different types of constraints were observed in CDRs, while FRs close to CDRs were sometimes primarily impacted. This study sheds light on the changes in flexibility in different regions of VHH that may impact their in silico design.
Keywords: Protein Blocks; antibody; disorder; flexibility; mobility; molecular dynamics simulation; nanobody; single-chain antibody; structural alphabet; sybody.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures


Similar articles
-
Global analysis of VHHs framework regions with a structural alphabet.Biochimie. 2016 Dec;131:11-19. doi: 10.1016/j.biochi.2016.09.005. Epub 2016 Sep 6. Biochimie. 2016. PMID: 27613403
-
Insights into Comparative Modeling of VHH Domains.Int J Mol Sci. 2021 Sep 9;22(18):9771. doi: 10.3390/ijms22189771. Int J Mol Sci. 2021. PMID: 34575931 Free PMC article.
-
Solution structure and backbone dynamics of an antigen-free heavy chain variable domain (VHH) from Llama.Proteins. 2002 Jun 1;47(4):546-55. doi: 10.1002/prot.10096. Proteins. 2002. PMID: 12001233
-
Single domain camel antibodies: current status.J Biotechnol. 2001 Jun;74(4):277-302. doi: 10.1016/s1389-0352(01)00021-6. J Biotechnol. 2001. PMID: 11526908 Review.
-
VHH Structural Modelling Approaches: A Critical Review.Int J Mol Sci. 2022 Mar 28;23(7):3721. doi: 10.3390/ijms23073721. Int J Mol Sci. 2022. PMID: 35409081 Free PMC article. Review.
Cited by
-
A Simple Analysis of the Second (Extra) Disulfide Bridge of VHHs.Molecules. 2024 Oct 14;29(20):4863. doi: 10.3390/molecules29204863. Molecules. 2024. PMID: 39459230 Free PMC article.
-
Evaluation of the Potential Impact of In Silico Humanization on VHH Dynamics.Int J Mol Sci. 2023 Sep 26;24(19):14586. doi: 10.3390/ijms241914586. Int J Mol Sci. 2023. PMID: 37834033 Free PMC article.
References
-
- Wrapp D., De Vlieger D., Corbett K.S., Torres G.M., Wang N., Van Breedam W., Roose K., van Schie L., Hoffmann M., Pöhlmann S., et al. Structural basis for potent neutralization of betacoronaviruses by single-domain camelid antibodies. Cell. 2020;181:1004–1015.e15. doi: 10.1016/j.cell.2020.04.031. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- S3D VHH program, N° SYNERGIE RE0022962/POE FEDER 2014-20 of the Conseil Régional de La Réunion - EU-H2020 and Université de la Réunion
- ANR-18-IDEX-0001/Agence Nationale de la Recherche
- ANR-11-LABX-0051/Agence Nationale de la Recherche
- ANR-11-IDEX-0005-02/Agence Nationale de la Recherche
- ANR-19-CE17-0021/Agence Nationale de la Recherche
LinkOut - more resources
Full Text Sources

