Development of an immune-related gene prognostic risk model and identification of an immune infiltration signature in the tumor microenvironment of colon cancer
- PMID: 36890467
- PMCID: PMC9996977
- DOI: 10.1186/s12876-023-02679-6
Development of an immune-related gene prognostic risk model and identification of an immune infiltration signature in the tumor microenvironment of colon cancer
Abstract
Background: Colon cancer is a common and highly malignant tumor. Its incidence is increasing rapidly with poor prognosis. At present, immunotherapy is a rapidly developing treatment for colon cancer. The aim of this study was to construct a prognostic risk model based on immune genes for early diagnosis and accurate prognostic prediction of colon cancer.
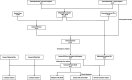
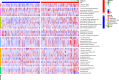
Methods: Transcriptome data and clinical data were downloaded from the cancer Genome Atlas database. Immunity genes were obtained from ImmPort database. The differentially expressed transcription factors (TFs) were obtained from Cistrome database. Differentially expressed (DE) immune genes were identified in 473 cases of colon cancer and 41 cases of normal adjacent tissues. An immune-related prognostic model of colon cancer was established and its clinical applicability was verified. Among 318 tumor-related transcription factors, differentially expressed transcription factors were finally obtained, and a regulatory network was constructed according to the up-down regulatory relationship.
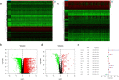
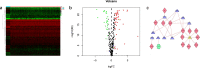
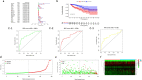
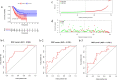
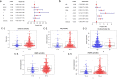
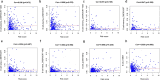
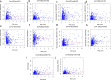
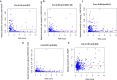
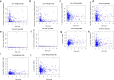
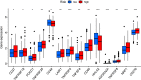
Results: A total of 477 DE immune genes (180 up-regulated and 297 down-regulated) were detected. We developed and validated twelve immune gene models for colon cancer, including SLC10A2, FABP4, FGF2, CCL28, IGKV1-6, IGLV6-57, ESM1, UCN, UTS2, VIP, IL1RL2, NGFR. The model was proved to be an independent prognostic variable with good prognostic ability. A total of 68 DE TFs (40 up-regulated and 23 down-regulated) were obtained. The regulation network between TF and immune genes was plotted by using TF as source node and immune genes as target node. In addition, Macrophage, Myeloid Dendritic cell and CD4+ T cell increased with the increase of risk score.
Conclusion: We developed and validated twelve immune gene models for colon cancer, including SLC10A2, FABP4, FGF2, CCL28, IGKV1-6, IGLV6-57, ESM1, UCN, UTS2, VIP, IL1RL2, NGFR. This model can be used as a tool variable to predict the prognosis of colon cancer.
Keywords: Colon cancer; Immune gene; Prognosis; Risk model.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Similar articles
-
Development and validation of a five-immune gene prognostic risk model in colon cancer.BMC Cancer. 2020 May 6;20(1):395. doi: 10.1186/s12885-020-06799-0. BMC Cancer. 2020. PMID: 32375704 Free PMC article.
-
A Prognostic Model Based on the Immune-related Genes in Colon Adenocarcinoma.Int J Med Sci. 2020 Jul 19;17(13):1879-1896. doi: 10.7150/ijms.45813. eCollection 2020. Int J Med Sci. 2020. PMID: 32788867 Free PMC article.
-
Identification prognosis-associated immune genes in colon adenocarcinoma.Biosci Rep. 2020 Nov 27;40(11):BSR20201734. doi: 10.1042/BSR20201734. Biosci Rep. 2020. PMID: 33140821 Free PMC article.
-
Prognostic Model of Colorectal Cancer Constructed by Eight Immune-Related Genes.Front Mol Biosci. 2020 Nov 27;7:604252. doi: 10.3389/fmolb.2020.604252. eCollection 2020. Front Mol Biosci. 2020. PMID: 33330631 Free PMC article.
-
Establishment and validation of an aging-related risk signature associated with prognosis and tumor immune microenvironment in breast cancer.Eur J Med Res. 2022 Dec 29;27(1):317. doi: 10.1186/s40001-022-00924-4. Eur J Med Res. 2022. PMID: 36581948 Free PMC article.
Cited by
-
IL1RL2 is related to the expression and prognosis of bladder cancer.Mol Clin Oncol. 2024 Aug 13;21(4):75. doi: 10.3892/mco.2024.2773. eCollection 2024 Oct. Mol Clin Oncol. 2024. PMID: 39170626 Free PMC article.
References
-
- Hossain MS, Karuniawati H, Jairoun AA, Urbi Z, Ooi DJ, John A, Lim YC, Kibria KMK, Mohiuddin AKM, Ming LC, et al. Colorectal cancer: a review of carcinogenesis, global epidemiology, current challenges, risk factors, preventive and treatment strategies. Cancers. 2022;14(7):1732. doi: 10.3390/cancers14071732. - DOI - PMC - PubMed
-
- Schmoll HJ, Van Cutsem E, Stein A, Valentini V, Glimelius B, Haustermans K, Nordlinger B, van de Velde CJ, Balmana J, Regula J, et al. ESMO consensus guidelines for management of patients with colon and rectal cancer. A personalized approach to clinical decision making. Ann Oncol. 2012;23(10):2479–2516. doi: 10.1093/annonc/mds236. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

