The structure, function, and pharmacology of MRGPRs
- PMID: 36870785
- PMCID: PMC10066734
- DOI: 10.1016/j.tips.2023.02.002
The structure, function, and pharmacology of MRGPRs
Abstract
Mas-related G protein-coupled receptor (MRGPR) family members play important roles in the sensation of noxious stimuli and represent novel targets for the treatment of itch and pain. MRGPRs recognize a diversity of agonists and display complicated downstream signaling profiles, high sequence diversity across species, and many polymorphisms in humans. The recent structural advances on MRGPRs reveal unique structural features and diverse agonist recognition modes of this receptor family, which should facilitate the structure-based drug discovery at MRGPRs. In addition, the newly discovered ligands also provide valuable tools to explore the function and the therapeutic potential of MRGPRs. In this review, we discuss these progresses in our understanding of MRGPRs and highlight the challenges and potential opportunities for the future drug discovery at these receptors.
Keywords: MRGPRs; drug discovery; function; ligand recognition; polymorphism; structure.
Copyright © 2023 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of interests B.L.R. and C.C. are listed as inventors on a patent application related to MRGPRX2 antagonists.
Figures
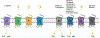


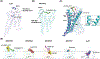


Similar articles
-
Beyond somatosensation: Mrgprs in mucosal tissues.Neurosci Lett. 2021 Mar 23;748:135689. doi: 10.1016/j.neulet.2021.135689. Epub 2021 Feb 11. Neurosci Lett. 2021. PMID: 33582191 Review.
-
MAS and its related G protein-coupled receptors, Mrgprs.Pharmacol Rev. 2014 Oct;66(4):1080-105. doi: 10.1124/pr.113.008136. Pharmacol Rev. 2014. PMID: 25244929 Review.
-
mMrgprA3/mMrgprC11/hMrgprX1: Potential therapeutic targets for allergic contact dermatitis-induced pruritus in mice and humans.Contact Dermatitis. 2022 Apr;86(4):286-294. doi: 10.1111/cod.14051. Epub 2022 Feb 8. Contact Dermatitis. 2022. PMID: 35066892
-
The role of the Mrgpr receptor family in itch.Handb Exp Pharmacol. 2015;226:71-88. doi: 10.1007/978-3-662-44605-8_5. Handb Exp Pharmacol. 2015. PMID: 25861775 Review.
-
Sensory neuron-specific GPCR Mrgprs are itch receptors mediating chloroquine-induced pruritus.Cell. 2009 Dec 24;139(7):1353-65. doi: 10.1016/j.cell.2009.11.034. Epub 2009 Dec 10. Cell. 2009. PMID: 20004959 Free PMC article.
Cited by
-
Orphan G Protein-Coupled Receptor GPR37 as an Emerging Therapeutic Target.ACS Chem Neurosci. 2023 Sep 20;14(18):3318-3334. doi: 10.1021/acschemneuro.3c00479. Epub 2023 Sep 7. ACS Chem Neurosci. 2023. PMID: 37676000 Free PMC article. Review.
-
Molecular basis of opioid receptor signaling.Cell. 2023 Nov 22;186(24):5203-5219. doi: 10.1016/j.cell.2023.10.029. Cell. 2023. PMID: 37995655 Free PMC article. Review.
-
Mrgprb2-mediated mast cell activation exacerbates Modic changes by regulating immune niches.Exp Mol Med. 2024 May;56(5):1178-1192. doi: 10.1038/s12276-024-01230-1. Epub 2024 May 1. Exp Mol Med. 2024. PMID: 38689089 Free PMC article.
-
A Skin Testing Strategy for Non-IgE-Mediated Reactions Associated With Vancomycin.J Allergy Clin Immunol Pract. 2024 Nov;12(11):3025-3033.e6. doi: 10.1016/j.jaip.2024.07.028. Epub 2024 Aug 6. J Allergy Clin Immunol Pract. 2024. PMID: 39117269
-
Modulation of the Mas-Related G Protein-Coupled Receptor X2 (MRGPRX2) by Xenobiotic Compounds and Its Relevance to Human Diseases.J Xenobiot. 2024 Mar 13;14(1):380-403. doi: 10.3390/jox14010024. J Xenobiot. 2024. PMID: 38535499 Free PMC article. Review.
References
-
- Young D et al. (1986) Isolation and characterization of a new cellular oncogene encoding a protein with multiple potential transmembrane domains. Cell 45 (5), 711–9. - PubMed
-
- Dong X et al. (2001) A diverse family of GPCRs expressed in specific subsets of nociceptive sensory neurons. Cell 106 (5), 619–32. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

