This is a preprint.
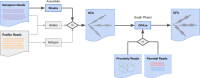
Phased nanopore assembly with Shasta and modular graph phasing with GFAse
- PMID: 36865218
- PMCID: PMC9980101
- DOI: 10.1101/2023.02.21.529152
Phased nanopore assembly with Shasta and modular graph phasing with GFAse
Update in
-
Phased nanopore assembly with Shasta and modular graph phasing with GFAse.Genome Res. 2024 Apr 25;34(3):454-468. doi: 10.1101/gr.278268.123. Genome Res. 2024. PMID: 38627094 Free PMC article.
Abstract
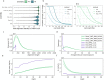
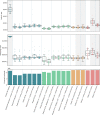
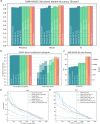
As a step towards simplifying and reducing the cost of haplotype resolved de novo assembly, we describe new methods for accurately phasing nanopore data with the Shasta genome assembler and a modular tool for extending phasing to the chromosome scale called GFAse. We test using new variants of Oxford Nanopore Technologies' (ONT) PromethION sequencing, including those using proximity ligation and show that newer, higher accuracy ONT reads substantially improve assembly quality.
Figures

Similar articles
-
Phased nanopore assembly with Shasta and modular graph phasing with GFAse.Genome Res. 2024 Apr 25;34(3):454-468. doi: 10.1101/gr.278268.123. Genome Res. 2024. PMID: 38627094 Free PMC article.
-
Chromosome-scale assembly comparison of the Korean Reference Genome KOREF from PromethION and PacBio with Hi-C mapping information.Gigascience. 2019 Dec 1;8(12):giz125. doi: 10.1093/gigascience/giz125. Gigascience. 2019. PMID: 31794015 Free PMC article.
-
Benchmarking Long-Read Assemblers for Genomic Analyses of Bacterial Pathogens Using Oxford Nanopore Sequencing.Int J Mol Sci. 2020 Dec 1;21(23):9161. doi: 10.3390/ijms21239161. Int J Mol Sci. 2020. PMID: 33271875 Free PMC article.
-
Haplotyping-Assisted Diploid Assembly and Variant Detection with Linked Reads.Methods Mol Biol. 2023;2590:161-182. doi: 10.1007/978-1-0716-2819-5_11. Methods Mol Biol. 2023. PMID: 36335499 Review.
-
Oxford Nanopore MinION Sequencing and Genome Assembly.Genomics Proteomics Bioinformatics. 2016 Oct;14(5):265-279. doi: 10.1016/j.gpb.2016.05.004. Epub 2016 Sep 17. Genomics Proteomics Bioinformatics. 2016. PMID: 27646134 Free PMC article. Review.
References
-
- Marchini J. & Howie B. Genotype imputation for genome-wide association studies. Nat. Rev. Genet. 11, 499–511 (2010). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
