The RNA binding protein DND1 is elevated in a subpopulation of pro-spermatogonia and targets chromatin modifiers and translational machinery during late gestation
- PMID: 36857387
- PMCID: PMC10010562
- DOI: 10.1371/journal.pgen.1010656
The RNA binding protein DND1 is elevated in a subpopulation of pro-spermatogonia and targets chromatin modifiers and translational machinery during late gestation
Abstract
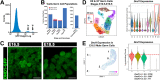
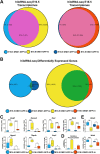
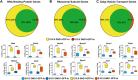
DND1 is essential to maintain germ cell identity. Loss of Dnd1 function results in germ cell differentiation to teratomas in some inbred strains of mice or to somatic fates in zebrafish. Using our knock-in mouse line in which a functional fusion protein between DND1 and GFP is expressed from the endogenous locus (Dnd1GFP), we distinguished two male germ cell (MGC) populations during late gestation cell cycle arrest (G0), consistent with recent reports of heterogeneity among MGCs. Most MGCs express lower levels of DND1-GFP (DND1-GFP-lo), but some MGCs express elevated levels of DND1-GFP (DND1-GFP-hi). A RNA-seq time course confirmed high Dnd1 transcript levels in DND1-GFP-hi cells along with 5-10-fold higher levels for multiple epigenetic regulators. Using antibodies against DND1-GFP for RNA immunoprecipitation (RIP)-sequencing, we identified multiple epigenetic and translational regulators that are binding targets of DND1 during G0 including DNA methyltransferases (Dnmts), histone deacetylases (Hdacs), Tudor domain proteins (Tdrds), actin dependent regulators (Smarcs), and a group of ribosomal and Golgi proteins. These data suggest that in DND1-GFP-hi cells, DND1 hosts coordinating mRNA regulons that consist of functionally related and localized groups of epigenetic enzymes and translational components.
Copyright: © 2023 Ruthig et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have no competing interests to disclose.
Figures



Similar articles
-
The RNA-binding protein DND1 acts sequentially as a negative regulator of pluripotency and a positive regulator of epigenetic modifiers required for germ cell reprogramming.Development. 2019 Jul 25;146(19):dev175950. doi: 10.1242/dev.175950. Development. 2019. PMID: 31253634 Free PMC article.
-
A transgenic DND1GFP fusion allele reports in vivo expression and RNA-binding targets in undifferentiated mouse germ cells†.Biol Reprod. 2021 Apr 1;104(4):861-874. doi: 10.1093/biolre/ioaa233. Biol Reprod. 2021. PMID: 33394034 Free PMC article.
-
Regulation of male germ cell cycle arrest and differentiation by DND1 is modulated by genetic background.Development. 2011 Jan;138(1):23-32. doi: 10.1242/dev.057000. Epub 2010 Nov 29. Development. 2011. PMID: 21115610 Free PMC article.
-
The Role of DND1 in Cancers.Cancers (Basel). 2021 Jul 22;13(15):3679. doi: 10.3390/cancers13153679. Cancers (Basel). 2021. PMID: 34359581 Free PMC article. Review.
-
Interplay of RNA-binding proteins controls germ cell development in zebrafish.J Genet Genomics. 2024 Sep;51(9):889-899. doi: 10.1016/j.jgg.2024.06.020. Epub 2024 Jul 4. J Genet Genomics. 2024. PMID: 38969260 Review.
References
-
- Sakurai T, Iguchi T, Moriwaki K, Noguchi M. The ter mutation first causes primordial germ cell deficiency in ter/ter mouse embryos at 8 days of gestation. Development, Growth & Differentiation. 1995;37(3):293–302. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

