SARS-CoV-2 Omicron Subvariants Balance Host Cell Membrane, Receptor, and Antibody Docking via an Overlapping Target Site
- PMID: 36851661
- PMCID: PMC9967007
- DOI: 10.3390/v15020447
SARS-CoV-2 Omicron Subvariants Balance Host Cell Membrane, Receptor, and Antibody Docking via an Overlapping Target Site
Abstract
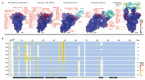
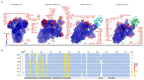
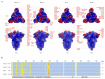
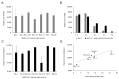
Variants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) are emerging rapidly and offer surfaces that are optimized for recognition of host cell membranes while also evading antibodies arising from vaccinations and previous infections. Host cell infection is a multi-step process in which spike heads engage lipid bilayers and one or more angiotensin-converting enzyme 2 (ACE-2) receptors. Here, the membrane binding surfaces of Omicron subvariants are compared using cryo-electron microscopy (cEM) structures of spike trimers from BA.2, BA.2.12.1, BA.2.13, BA.2.75, BA.3, BA.4, and BA.5 viruses. Despite significant differences around mutated sites, they all maintain strong membrane binding propensities that first appeared in BA.1. Both their closed and open states retain elevated membrane docking capacities, although the presence of more closed than open states diminishes opportunities to bind receptors while enhancing membrane engagement. The electrostatic dipoles are generally conserved. However, the BA.2.75 spike dipole is compromised, and its ACE-2 affinity is increased, and BA.3 exhibits the opposite pattern. We propose that balancing the functional imperatives of a stable, readily cleavable spike that engages both lipid bilayers and receptors while avoiding host defenses underlies betacoronavirus evolution. This provides predictive criteria for rationalizing future pandemic waves and COVID-19 transmissibility while illuminating critical sites and strategies for simultaneously combating multiple variants.
Keywords: MODA; SARS-CoV-2; coronavirus; delta; lipid bilayer; membrane docking; omicron; spike protein; variants.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
The evolution of the spike protein and hACE2 interface of SARS-CoV-2 omicron variants determined by hydrogen bond formation.Brief Funct Genomics. 2023 May 18;22(3):291-301. doi: 10.1093/bfgp/elac053. Brief Funct Genomics. 2023. PMID: 36723978
-
Exploring Conformational Landscapes and Cryptic Binding Pockets in Distinct Functional States of the SARS-CoV-2 Omicron BA.1 and BA.2 Trimers: Mutation-Induced Modulation of Protein Dynamics and Network-Guided Prediction of Variant-Specific Allosteric Binding Sites.Viruses. 2023 Sep 27;15(10):2009. doi: 10.3390/v15102009. Viruses. 2023. PMID: 37896786 Free PMC article.
-
In silico study of SARS-CoV-2 spike protein RBD and human ACE-2 affinity dynamics across variants and Omicron subvariants.J Med Virol. 2023 Jan;95(1):e28406. doi: 10.1002/jmv.28406. J Med Virol. 2023. PMID: 36519577 Free PMC article.
-
Physicochemical effects of emerging exchanges on the spike protein's RBM of the SARS-CoV-2 Omicron subvariants BA.1-BA.5 and its influence on the biological properties and attributes developed by these subvariants.Virology. 2023 Oct;587:109850. doi: 10.1016/j.virol.2023.109850. Epub 2023 Jul 28. Virology. 2023. PMID: 37562286 Review.
-
The emergence of SARS-CoV-2 Omicron subvariants: current situation and future trends.Infez Med. 2022 Dec 1;30(4):480-494. doi: 10.53854/liim-3004-2. eCollection 2022. Infez Med. 2022. PMID: 36482957 Free PMC article. Review.
Cited by
-
Exploring the Syndecan-Mediated Cellular Internalization of the SARS-CoV-2 Omicron Variant.Int J Mol Sci. 2023 Sep 15;24(18):14140. doi: 10.3390/ijms241814140. Int J Mol Sci. 2023. PMID: 37762442 Free PMC article.
References
-
- Satturwar S., Fowkes M., Farver C., Wilson A.M., Eccher A., Girolami I., Pujadas E., Bryce C., Salem F., El Jamal S.M., et al. Postmortem Findings Associated With SARS-CoV-2: Systematic Review and Meta-analysis. Am. J. Surg. Pathol. 2021;45:587–603. doi: 10.1097/PAS.0000000000001650. - DOI - PMC - PubMed
-
- Arora P., Nehlmeier I., Kempf A., Cossmann A., Schulz S.R., Dopfer-Jablonka A., Baier E., Tampe B., Moerer O., Dickel S., et al. Lung cell entry, cell-cell fusion capacity, and neutralisation sensitivity of omicron sublineage BA.2.75. Lancet Infect. Dis. 2022;22:1537–1538. doi: 10.1016/S1473-3099(22)00591-6. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

