Phase Separation: The Robust Modulator of Innate Antiviral Signaling and SARS-CoV-2 Infection
- PMID: 36839515
- PMCID: PMC9962166
- DOI: 10.3390/pathogens12020243
Phase Separation: The Robust Modulator of Innate Antiviral Signaling and SARS-CoV-2 Infection
Abstract
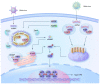
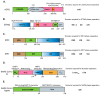
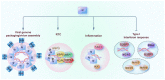
SARS-CoV-2 has been a pandemic threat to human health and the worldwide economy, but efficient treatments are still lacking. Type I and III interferons are essential for controlling viral infection, indicating that antiviral innate immune signaling is critical for defense against viral infection. Phase separation, one of the basic molecular processes, governs multiple cellular activities, such as cancer progression, microbial infection, and signaling transduction. Notably, recent studies suggest that phase separation regulates antiviral signaling such as the RLR and cGAS-STING pathways. Moreover, proper phase separation of viral proteins is essential for viral replication and pathogenesis. These observations indicate that phase separation is a critical checkpoint for virus and host interaction. In this study, we summarize the recent advances concerning the regulation of antiviral innate immune signaling and SARS-CoV-2 infection by phase separation. Our review highlights the emerging notion that phase separation is the robust modulator of innate antiviral signaling and viral infection.
Keywords: N protein; NSP8; RLR; SARS-CoV-2; cGAS–STING; phase separation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Regulation of SARS-CoV-2 infection and antiviral innate immunity by ubiquitination and ubiquitin-like conjugation.Biochim Biophys Acta Gene Regul Mech. 2023 Dec;1866(4):194984. doi: 10.1016/j.bbagrm.2023.194984. Epub 2023 Sep 17. Biochim Biophys Acta Gene Regul Mech. 2023. PMID: 37717938 Review.
-
SARS-CoV-2 ORF3a inhibits cGAS-STING-mediated autophagy flux and antiviral function.J Med Virol. 2023 Jan;95(1):e28175. doi: 10.1002/jmv.28175. Epub 2022 Oct 8. J Med Virol. 2023. PMID: 36163413 Free PMC article.
-
SARS-CoV-2 ORF10 antagonizes STING-dependent interferon activation and autophagy.J Med Virol. 2022 Nov;94(11):5174-5188. doi: 10.1002/jmv.27965. Epub 2022 Jul 7. J Med Virol. 2022. PMID: 35765167 Free PMC article.
-
Progress of cGAS-STING signaling in response to SARS-CoV-2 infection.Front Immunol. 2022 Dec 7;13:1010911. doi: 10.3389/fimmu.2022.1010911. eCollection 2022. Front Immunol. 2022. PMID: 36569852 Free PMC article. Review.
-
Sensing of cytoplasmic chromatin by cGAS activates innate immune response in SARS-CoV-2 infection.Signal Transduct Target Ther. 2021 Nov 3;6(1):382. doi: 10.1038/s41392-021-00800-3. Signal Transduct Target Ther. 2021. PMID: 34732709 Free PMC article.
Cited by
-
Regulation of cellular senescence by innate immunity.Biophys Rep. 2023 Dec 31;9(6):338-351. doi: 10.52601/bpr.2023.230032. Biophys Rep. 2023. PMID: 38524701 Free PMC article.
-
Host mitochondria: more than an organelle in SARS-CoV-2 infection.Front Cell Infect Microbiol. 2023 Aug 25;13:1228275. doi: 10.3389/fcimb.2023.1228275. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37692170 Free PMC article. Review.
-
Intracellular Degradation of SARS-CoV-2 N-Protein Caused by Modular Nanotransporters Containing Anti-N-Protein Monobody and a Sequence That Recruits the Keap1 E3 Ligase.Pharmaceutics. 2023 Dec 19;16(1):4. doi: 10.3390/pharmaceutics16010004. Pharmaceutics. 2023. PMID: 38276482 Free PMC article.
-
A Tiny Viral Protein, SARS-CoV-2-ORF7b: Functional Molecular Mechanisms.Biomolecules. 2024 Apr 30;14(5):541. doi: 10.3390/biom14050541. Biomolecules. 2024. PMID: 38785948 Free PMC article.
References
-
- Zheng Y., Gao C. Fine-tuning of antiviral innate immunity by ubiquitination. Adv. Immunol. 2020;145:95–128. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

