Distinct Conformations of SARS-CoV-2 Omicron Spike Protein and Its Interaction with ACE2 and Antibody
- PMID: 36835186
- PMCID: PMC9967551
- DOI: 10.3390/ijms24043774
Distinct Conformations of SARS-CoV-2 Omicron Spike Protein and Its Interaction with ACE2 and Antibody
Abstract
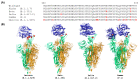
Since November 2021, Omicron has been the dominant severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variant that causes the coronavirus disease 2019 (COVID-19) and has continuously impacted human health. Omicron sublineages are still increasing and cause increased transmission and infection rates. The additional 15 mutations on the receptor binding domain (RBD) of Omicron spike proteins change the protein conformation, enabling the Omicron variant to evade neutralizing antibodies. For this reason, many efforts have been made to design new antigenic variants to induce effective antibodies in SARS-CoV-2 vaccine development. However, understanding the different states of Omicron spike proteins with and without external molecules has not yet been addressed. In this review, we analyze the structures of the spike protein in the presence and absence of angiotensin-converting enzyme 2 (ACE2) and antibodies. Compared to previously determined structures for the wildtype spike protein and other variants such as alpha, beta, delta, and gamma, the Omicron spike protein adopts a partially open form. The open-form spike protein with one RBD up is dominant, followed by the open-form spike protein with two RBD up, and the closed-form spike protein with the RBD down. It is suggested that the competition between antibodies and ACE2 induces interactions between adjacent RBDs of the spike protein, which lead to a partially open form of the Omicron spike protein. The comprehensive structural information of Omicron spike proteins could be helpful for the efficient design of vaccines against the Omicron variant.
Keywords: Omicron; SARS-CoV-2; angiotensin-converting enzyme 2; antibody; open-form spike protein; receptor binding domain.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

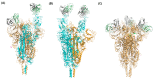
Similar articles
-
Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.Science. 2022 Mar 4;375(6584):1048-1053. doi: 10.1126/science.abn8863. Epub 2022 Feb 8. Science. 2022. PMID: 35133176 Free PMC article.
-
Structural basis and analysis of hamster ACE2 binding to different SARS-CoV-2 spike RBDs.J Virol. 2024 Mar 19;98(3):e0115723. doi: 10.1128/jvi.01157-23. Epub 2024 Feb 2. J Virol. 2024. PMID: 38305152 Free PMC article.
-
V367F Mutation in SARS-CoV-2 Spike RBD Emerging during the Early Transmission Phase Enhances Viral Infectivity through Increased Human ACE2 Receptor Binding Affinity.J Virol. 2021 Jul 26;95(16):e0061721. doi: 10.1128/JVI.00617-21. Epub 2021 Jul 26. J Virol. 2021. PMID: 34105996 Free PMC article.
-
The Biological Functions and Clinical Significance of SARS-CoV-2 Variants of Corcern.Front Med (Lausanne). 2022 May 20;9:849217. doi: 10.3389/fmed.2022.849217. eCollection 2022. Front Med (Lausanne). 2022. PMID: 35669924 Free PMC article. Review.
-
Structural basis of severe acute respiratory syndrome coronavirus 2 infection.Curr Opin HIV AIDS. 2021 Jan;16(1):74-81. doi: 10.1097/COH.0000000000000658. Curr Opin HIV AIDS. 2021. PMID: 33186231 Review.
Cited by
-
Comparative Computational Analysis of Spike Protein Structural Stability in SARS-CoV-2 Omicron Subvariants.Int J Mol Sci. 2023 Nov 8;24(22):16069. doi: 10.3390/ijms242216069. Int J Mol Sci. 2023. PMID: 38003257 Free PMC article.
-
Weak Value Amplification Based Optical Sensor for High Throughput Real-Time Immunoassay of SARS-CoV-2 Spike Protein.Biosensors (Basel). 2024 Jul 8;14(7):332. doi: 10.3390/bios14070332. Biosensors (Basel). 2024. PMID: 39056608 Free PMC article.
-
Epitopes of an antibody that neutralizes a wide range of SARS-CoV-2 variants in a conserved subdomain 1 of the spike protein.J Virol. 2024 May 14;98(5):e0041624. doi: 10.1128/jvi.00416-24. Epub 2024 Apr 16. J Virol. 2024. PMID: 38624232 Free PMC article.
-
Developing a SARS-CoV-2 main protease binding prediction random forest model for drug repurposing for COVID-19 treatment.Exp Biol Med (Maywood). 2023 Nov;248(21):1927-1936. doi: 10.1177/15353702231209413. Epub 2023 Nov 24. Exp Biol Med (Maywood). 2023. PMID: 37997891 Free PMC article.
-
Biophysical principles predict fitness of SARS-CoV-2 variants.bioRxiv [Preprint]. 2024 Jan 22:2023.07.23.549087. doi: 10.1101/2023.07.23.549087. bioRxiv. 2024. Update in: Proc Natl Acad Sci U S A. 2024 Jun 4;121(23):e2314518121. doi: 10.1073/pnas.2314518121. PMID: 37577536 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

