Plasma Proteomic Variables Related to COVID-19 Severity: An Untargeted nLC-MS/MS Investigation
- PMID: 36834989
- PMCID: PMC9962231
- DOI: 10.3390/ijms24043570
Plasma Proteomic Variables Related to COVID-19 Severity: An Untargeted nLC-MS/MS Investigation
Abstract
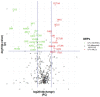
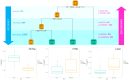
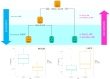
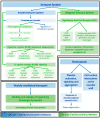
Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) infection leads to a wide range of clinical manifestations and determines the need for personalized and precision medicine. To better understand the biological determinants of this heterogeneity, we explored the plasma proteome of 43 COVID-19 patients with different outcomes by an untargeted liquid chromatography-mass spectrometry approach. The comparison between asymptomatic or pauci-symptomatic subjects (MILDs), and hospitalised patients in need of oxygen support therapy (SEVEREs) highlighted 29 proteins emerged as differentially expressed: 12 overexpressed in MILDs and 17 in SEVEREs. Moreover, a supervised analysis based on a decision-tree recognised three proteins (Fetuin-A, Ig lambda-2chain-C-region, Vitronectin) that are able to robustly discriminate between the two classes independently from the infection stage. In silico functional annotation of the 29 deregulated proteins pinpointed several functions possibly related to the severity; no pathway was associated exclusively to MILDs, while several only to SEVEREs, and some associated to both MILDs and SEVEREs; SARS-CoV-2 signalling pathway was significantly enriched by proteins up-expressed in SEVEREs (SAA1/2, CRP, HP, LRG1) and in MILDs (GSN, HRG). In conclusion, our analysis could provide key information for 'proteomically' defining possible upstream mechanisms and mediators triggering or limiting the domino effect of the immune-related response and characterizing severe exacerbations.
Keywords: COVID-19; SARS-CoV-2; blood; mass spectrometry; plasma; proteomics; severe.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Molecular fingerprint by omics-based approaches in saliva from patients affected by SARS-CoV-2 infection.J Mass Spectrom. 2024 Sep;59(9):e5082. doi: 10.1002/jms.5082. J Mass Spectrom. 2024. PMID: 39228271
-
Salivary proteomic analysis in asymptomatic and symptomatic SARS-CoV-2 infection: Innate immunity, taste perception and FABP5 proteins make the difference.Clin Chim Acta. 2022 Dec 1;537:26-37. doi: 10.1016/j.cca.2022.09.023. Epub 2022 Oct 10. Clin Chim Acta. 2022. PMID: 36228679 Free PMC article.
-
Quantitative proteomics of hamster lung tissues infected with SARS-CoV-2 reveal host factors having implication in the disease pathogenesis and severity.FASEB J. 2021 Jul;35(7):e21713. doi: 10.1096/fj.202100431R. FASEB J. 2021. PMID: 34105201 Free PMC article.
-
[Applications of separation technology in novel coronavirus research, epidemic prevention and detection].Se Pu. 2021 Jul 8;39(7):679-685. doi: 10.3724/SP.J.1123.2021.03022. Se Pu. 2021. PMID: 34227364 Free PMC article. Review. Chinese.
-
Mass spectrometry-based proteomic platforms for better understanding of SARS-CoV-2 induced pathogenesis and potential diagnostic approaches.Proteomics. 2021 May;21(10):e2000279. doi: 10.1002/pmic.202000279. Epub 2021 May 5. Proteomics. 2021. PMID: 33860983 Free PMC article. Review.
Cited by
-
Alterations in plasma proteome during acute COVID-19 and recovery.Mol Med. 2024 Aug 25;30(1):131. doi: 10.1186/s10020-024-00898-5. Mol Med. 2024. PMID: 39183264 Free PMC article.
-
The disruptive role of LRG1 on the vasculature and perivascular microenvironment.Front Cardiovasc Med. 2024 Apr 30;11:1386177. doi: 10.3389/fcvm.2024.1386177. eCollection 2024. Front Cardiovasc Med. 2024. PMID: 38745756 Free PMC article. Review.
-
Plasma Proteins Associated with COVID-19 Severity in Puerto Rico.Int J Mol Sci. 2024 May 16;25(10):5426. doi: 10.3390/ijms25105426. Int J Mol Sci. 2024. PMID: 38791465 Free PMC article.
-
Towards the Definition of the Molecular Hallmarks of Idiopathic Membranous Nephropathy in Serum Proteome: A DIA-PASEF Approach.Int J Mol Sci. 2023 Jul 21;24(14):11756. doi: 10.3390/ijms241411756. Int J Mol Sci. 2023. PMID: 37511514 Free PMC article.
-
Activation of the MAPK network provides a survival advantage during the course of COVID-19-induced sepsis: a real-world evidence analysis of a multicenter COVID-19 Sepsis Cohort.Infection. 2025 Feb;53(1):107-115. doi: 10.1007/s15010-024-02325-7. Epub 2024 Jun 19. Infection. 2025. PMID: 38896372 Free PMC article.
References
-
- Ciccosanti F., Antonioli M., Sacchi A., Notari S., Farina A., Beccacece A., Fusto M., Vergori A., D’Offizi G., Taglietti F., et al. Proteomic analysis identifies a signature of disease severity in the plasma of COVID-19 pneumonia patients associated to neutrophil, platelet and complement activation. Clin. Proteom. 2022;19:38. doi: 10.1186/s12014-022-09377-7. - DOI - PMC - PubMed
MeSH terms
Grants and funding
- FAR 2017-2021/FAR 2017-2021
- Pupa Ferrari Onlus/Fondazione Gigi & Pupa Ferrari Onlus
- regional law n° 9/2020, resolution n° 3776/2020"/Regione Lombardia, regional law n° 9/2020, resolution n° 3776/2020": Programma degli interventi per la ripresa economica: sviluppo di nuovi accordi di collaborazione con le università per la Ricerca, l'Innovazione e il Trasferimento tecnologico: NephropaT
- POR FESR 2014-2020/Regione Lombardia POR FESR 2014-2020. Misura a sostegno dello sviluppo di collaborazioni per l'identificazione di terapie e sistemi di diagnostica, protezione e analisi per contrastare l'emergenza Coronavirus e altre emergenze virali del futuro: Caratteri
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

