circINSR Inhibits Adipogenic Differentiation of Adipose-Derived Stromal Vascular Fractions through the miR-152/ MEOX2 Axis in Sheep
- PMID: 36834919
- PMCID: PMC9964708
- DOI: 10.3390/ijms24043501
circINSR Inhibits Adipogenic Differentiation of Adipose-Derived Stromal Vascular Fractions through the miR-152/ MEOX2 Axis in Sheep
Abstract
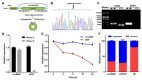
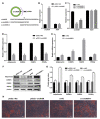
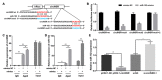
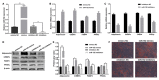
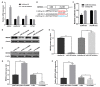
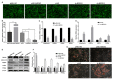
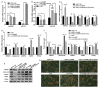
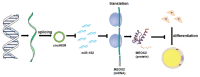
Adipose tissue plays a crucial role in energy metabolism. Several studies have shown that circular RNA (circRNA) is involved in the regulation of fat development and lipid metabolism. However, little is known about their involvement in the adipogenic differentiation of ovine stromal vascular fractions (SVFs). Here, based on previous sequencing data and bioinformatics analysis, a novel circINSR was identified in sheep, which acts as a sponge to promote miR-152 in inhibiting the adipogenic differentiation of ovine SVFs. The interactions between circINSR and miR-152 were examined using bioinformatics, luciferase assays, and RNA immunoprecipitation. Of note, we found that circINSR was involved in adipogenic differentiation via the miR-152/mesenchyme homeobox 2 (MEOX2) pathway. MEOX2 inhibited adipogenic differentiation of ovine SVFs and miR-152 inhibited the expression of MEOX2. In other words, circINSR directly isolates miR-152 in the cytoplasm and inhibits its ability to promote adipogenic differentiation of ovine SVFs. In summary, this study revealed the role of circINSR in the adipogenic differentiation of ovine SVFs and its regulatory mechanisms, providing a reference for further interpretation of the development of ovine fat and its regulatory mechanisms.
Keywords: MEOX2; adipogenesis; circINSR; miR-152; sheep; stromal vascular fraction.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
miR-301a inhibits adipogenic differentiation of adipose-derived stromal vascular fractions by targeting HOXC8 in sheep.Anim Sci J. 2021 Jan-Dec;92(1):e13661. doi: 10.1111/asj.13661. Anim Sci J. 2021. PMID: 34856652
-
miR-136 Regulates the Proliferation and Adipogenic Differentiation of Adipose-Derived Stromal Vascular Fractions by Targeting HSD17B12.Int J Mol Sci. 2023 Oct 4;24(19):14892. doi: 10.3390/ijms241914892. Int J Mol Sci. 2023. PMID: 37834341 Free PMC article.
-
MiR-33a inhibits the adipogenic differentiation of ovine adipose-derived stromal vascular fraction cells by targeting SIRT6.Domest Anim Endocrinol. 2021 Jan;74:106513. doi: 10.1016/j.domaniend.2020.106513. Epub 2020 Jun 18. Domest Anim Endocrinol. 2021. PMID: 32653737
-
MiR-128-1-5p regulates differentiation of ovine stromal vascular fraction by targeting the KLF11 5'-UTR.Domest Anim Endocrinol. 2022 Jul;80:106711. doi: 10.1016/j.domaniend.2022.106711. Epub 2022 Feb 5. Domest Anim Endocrinol. 2022. PMID: 35338828
-
Adipogenic miRNA and meta-signature miRNAs involved in human adipocyte differentiation and obesity.Oncotarget. 2016 Jun 28;7(26):40830-40845. doi: 10.18632/oncotarget.8518. Oncotarget. 2016. PMID: 27049726 Free PMC article. Review.
References
-
- Pan Y., Jing J., Qiao L., Liu J., An L., Li B., Ren D., Liu W. MiRNA-seq reveals that miR-124-3p inhibits adipogenic differentiation of the stromal vascular fraction in sheep via targeting C/EBPα. Domest. Anim. Endocrinol. 2018;65:17–23. - PubMed
-
- He C., Zhang Q., Sun H., Cai R., Pang W. Role of miRNA and lncRNA in animal fat deposition-a review. Sheng Wu Gong Cheng Xue Bao Chin. J. Biotechnol. 2020;36:1504–1514. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

