Transfer RNA Modification Enzymes with a Thiouridine Synthetase, Methyltransferase and Pseudouridine Synthase (THUMP) Domain and the Nucleosides They Produce in tRNA
- PMID: 36833309
- PMCID: PMC9957541
- DOI: 10.3390/genes14020382
Transfer RNA Modification Enzymes with a Thiouridine Synthetase, Methyltransferase and Pseudouridine Synthase (THUMP) Domain and the Nucleosides They Produce in tRNA
Abstract
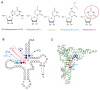
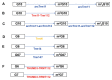
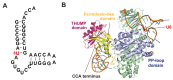
The existence of the thiouridine synthetase, methyltransferase and pseudouridine synthase (THUMP) domain was originally predicted by a bioinformatic study. Since the prediction of the THUMP domain more than two decades ago, many tRNA modification enzymes containing the THUMP domain have been identified. According to their enzymatic activity, THUMP-related tRNA modification enzymes can be classified into five types, namely 4-thiouridine synthetase, deaminase, methyltransferase, a partner protein of acetyltransferase and pseudouridine synthase. In this review, I focus on the functions and structures of these tRNA modification enzymes and the modified nucleosides they produce. Biochemical, biophysical and structural studies of tRNA 4-thiouridine synthetase, tRNA methyltransferases and tRNA deaminase have established the concept that the THUMP domain captures the 3'-end of RNA (in the case of tRNA, the CCA-terminus). However, in some cases, this concept is not simply applicable given the modification patterns observed in tRNA. Furthermore, THUMP-related proteins are involved in the maturation of other RNAs as well as tRNA. Moreover, the modified nucleosides, which are produced by the THUMP-related tRNA modification enzymes, are involved in numerous biological phenomena, and the defects of genes for human THUMP-related proteins are implicated in genetic diseases. In this review, these biological phenomena are also introduced.
Keywords: 4-thiouridine; C to U editing; N2-methylguanosine; N4-acetylcytidine; PUS10; deaminase; pseudouridine synthase; tRNA; tRNA methyltransferase; tRNA modification enzyme.
Conflict of interest statement
The author declares no conflict of interest.
Figures

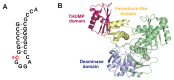

Similar articles
-
THUMP from archaeal tRNA:m22G10 methyltransferase, a genuine autonomously folding domain.Nucleic Acids Res. 2006 May 10;34(9):2483-94. doi: 10.1093/nar/gkl145. Print 2006. Nucleic Acids Res. 2006. PMID: 16687654 Free PMC article.
-
THUMP--a predicted RNA-binding domain shared by 4-thiouridine, pseudouridine synthases and RNA methylases.Trends Biochem Sci. 2001 Apr;26(4):215-7. doi: 10.1016/s0968-0004(01)01826-6. Trends Biochem Sci. 2001. PMID: 11295541
-
Crystal structure of a 4-thiouridine synthetase-RNA complex reveals specificity of tRNA U8 modification.Nucleic Acids Res. 2014 Jun;42(10):6673-85. doi: 10.1093/nar/gku249. Epub 2014 Apr 5. Nucleic Acids Res. 2014. PMID: 24705700 Free PMC article.
-
Transfer RNA methyltransferases with a SpoU-TrmD (SPOUT) fold and their modified nucleosides in tRNA.Biomolecules. 2017 Feb 28;7(1):23. doi: 10.3390/biom7010023. Biomolecules. 2017. PMID: 28264529 Free PMC article. Review.
-
[4Fe-4S]-dependent enzymes in non-redox tRNA thiolation.Biochim Biophys Acta Mol Cell Res. 2024 Oct;1871(7):119807. doi: 10.1016/j.bbamcr.2024.119807. Epub 2024 Aug 4. Biochim Biophys Acta Mol Cell Res. 2024. PMID: 39106920 Review.
Cited by
-
Deficiency of Acetyltransferase nat10 in Zebrafish Causes Developmental Defects in the Visual Function.Invest Ophthalmol Vis Sci. 2024 Feb 1;65(2):31. doi: 10.1167/iovs.65.2.31. Invest Ophthalmol Vis Sci. 2024. PMID: 38381411 Free PMC article.
-
PEGylated Dmoc phosphoramidites for sensitive oligodeoxynucleotide synthesis.Org Biomol Chem. 2023 Nov 22;21(45):9005-9010. doi: 10.1039/d3ob01495a. Org Biomol Chem. 2023. PMID: 37921008 Free PMC article.
-
RudS: bacterial desulfidase responsible for tRNA 4-thiouridine de-modification.Nucleic Acids Res. 2024 Sep 23;52(17):10543-10562. doi: 10.1093/nar/gkae716. Nucleic Acids Res. 2024. PMID: 39166491 Free PMC article.
-
Beyond the Anticodon: tRNA Core Modifications and Their Impact on Structure, Translation and Stress Adaptation.Genes (Basel). 2024 Mar 19;15(3):374. doi: 10.3390/genes15030374. Genes (Basel). 2024. PMID: 38540433 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

