Nonsense-Mediated mRNA Decay as a Mediator of Tumorigenesis
- PMID: 36833284
- PMCID: PMC9956241
- DOI: 10.3390/genes14020357
Nonsense-Mediated mRNA Decay as a Mediator of Tumorigenesis
Abstract
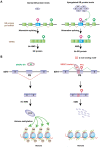
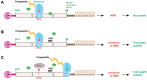
Nonsense-mediated mRNA decay (NMD) is an evolutionarily conserved and well-characterized biological mechanism that ensures the fidelity and regulation of gene expression. Initially, NMD was described as a cellular surveillance or quality control process to promote selective recognition and rapid degradation of erroneous transcripts harboring a premature translation-termination codon (PTC). As estimated, one-third of mutated and disease-causing mRNAs were reported to be targeted and degraded by NMD, suggesting the significance of this intricate mechanism in maintaining cellular integrity. It was later revealed that NMD also elicits down-regulation of many endogenous mRNAs without mutations (~10% of the human transcriptome). Therefore, NMD modulates gene expression to evade the generation of aberrant truncated proteins with detrimental functions, compromised activities, or dominant-negative effects, as well as by controlling the abundance of endogenous mRNAs. By regulating gene expression, NMD promotes diverse biological functions during development and differentiation, and facilitates cellular responses to adaptation, physiological changes, stresses, environmental insults, etc. Mutations or alterations (such as abnormal expression, degradation, post-translational modification, etc.) that impair the function or expression of proteins associated with the NMD pathway can be deleterious to cells and may cause pathological consequences, as implicated in developmental and intellectual disabilities, genetic defects, and cancer. Growing evidence in past decades has highlighted NMD as a critical driver of tumorigenesis. Advances in sequencing technologies provided the opportunity to identify many NMD substrate mRNAs in tumor samples compared to matched normal tissues. Interestingly, many of these changes are tumor-specific and are often fine-tuned in a tumor-specific manner, suggesting the complex regulation of NMD in cancer. Tumor cells differentially exploit NMD for survival benefits. Some tumors promote NMD to degrade a subset of mRNAs, such as those encoding tumor suppressors, stress response proteins, signaling proteins, RNA binding proteins, splicing factors, and immunogenic neoantigens. In contrast, some tumors suppress NMD to facilitate the expression of oncoproteins or other proteins beneficial for tumor growth and progression. In this review, we discuss how NMD is regulated as a critical mediator of oncogenesis to promote the development and progression of tumor cells. Understanding how NMD affects tumorigenesis differentially will pave the way for the development of more effective and less toxic, targeted therapeutic opportunities in the era of personalized medicine.
Keywords: cancer; gene expression; nonsense-mediated mRNA decay; splicing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Mechanisms and Regulation of Nonsense-Mediated mRNA Decay and Nonsense-Associated Altered Splicing in Lymphocytes.Int J Mol Sci. 2020 Feb 17;21(4):1335. doi: 10.3390/ijms21041335. Int J Mol Sci. 2020. PMID: 32079193 Free PMC article. Review.
-
Regulation of nonsense-mediated mRNA decay: implications for physiology and disease.Biochim Biophys Acta. 2013 Jun-Jul;1829(6-7):624-33. doi: 10.1016/j.bbagrm.2013.03.002. Epub 2013 Mar 13. Biochim Biophys Acta. 2013. PMID: 23500037 Free PMC article.
-
Nonsense-Mediated mRNA Decay in Development, Stress and Cancer.Adv Exp Med Biol. 2019;1157:41-83. doi: 10.1007/978-3-030-19966-1_3. Adv Exp Med Biol. 2019. PMID: 31342437
-
Nonsense-mediated mRNA decay in humans at a glance.J Cell Sci. 2016 Feb 1;129(3):461-7. doi: 10.1242/jcs.181008. Epub 2016 Jan 19. J Cell Sci. 2016. PMID: 26787741 Free PMC article. Review.
-
Perspective in Alternative Splicing Coupled to Nonsense-Mediated mRNA Decay.Int J Mol Sci. 2020 Dec 10;21(24):9424. doi: 10.3390/ijms21249424. Int J Mol Sci. 2020. PMID: 33321981 Free PMC article. Review.
Cited by
-
Caspases compromise SLU7 and UPF1 stability and NMD activity during hepatocarcinogenesis.JHEP Rep. 2024 May 9;6(8):101118. doi: 10.1016/j.jhepr.2024.101118. eCollection 2024 Aug. JHEP Rep. 2024. PMID: 39105183 Free PMC article.
-
Methamphetamine-induced region-specific transcriptomic and epigenetic changes in the brain of male rats.Commun Biol. 2023 Sep 27;6(1):991. doi: 10.1038/s42003-023-05355-3. Commun Biol. 2023. PMID: 37758941 Free PMC article.
-
The Art of Finding the Right Drug Target: Emerging Methods and Strategies.Pharmacol Rev. 2024 Aug 15;76(5):896-914. doi: 10.1124/pharmrev.123.001028. Pharmacol Rev. 2024. PMID: 38866560 Free PMC article. Review.
-
SR proteins in cancer: function, regulation, and small inhibitor.Cell Mol Biol Lett. 2024 May 22;29(1):78. doi: 10.1186/s11658-024-00594-6. Cell Mol Biol Lett. 2024. PMID: 38778254 Free PMC article. Review.
-
RNA Splicing in Cancer and Targeted Therapies.Genes (Basel). 2023 Oct 29;14(11):2020. doi: 10.3390/genes14112020. Genes (Basel). 2023. PMID: 38002963 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

