Emerging roles of the HECT E3 ubiquitin ligases in gastric cancer
- PMID: 36825281
- PMCID: PMC9941164
- DOI: 10.3389/pore.2023.1610931
Emerging roles of the HECT E3 ubiquitin ligases in gastric cancer
Abstract
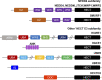
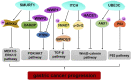
Gastric cancer (GC) is one of the most pernicious gastrointestinal tumors with extraordinarily high incidence and mortality. Ubiquitination modification of cellular signaling proteins has been shown to play important roles in GC tumorigenesis, progression, and prognosis. The E3 ubiquitin ligase is the crucial enzyme in the ubiquitination reaction and determines the specificity of ubiquitination substrates, and thus, the cellular effects. The HECT E3 ligases are the second largest E3 ubiquitin ligase family characterized by containing a HECT domain that has E3 ubiquitin ligase activity. The HECT E3 ubiquitin ligases have been found to engage in GC progression. However, whether HECT E3 ligases function as tumor promoters or tumor suppressors in GC remains controversial. In this review, we will focus on recent discoveries about the role of the HECT E3 ubiquitin ligases, especially members of the NEDD4 and other HECT E3 ligase subfamilies, in GC.
Keywords: HECT E3 ubiquitin ligases; gastric cancer; oncoprotein; tumor suppressor; ubiquitination.
Copyright © 2023 Sun, Tian, Chen, Yang and Lin.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Structural insights into a HECT-type E3 ligase AREL1 and its ubiquitination activities in vitro.J Biol Chem. 2019 Dec 27;294(52):19934-19949. doi: 10.1074/jbc.RA119.010327. Epub 2019 Nov 15. J Biol Chem. 2019. PMID: 31732561 Free PMC article.
-
The role of NEDD4 related HECT-type E3 ubiquitin ligases in defective autophagy in cancer cells: molecular mechanisms and therapeutic perspectives.Mol Med. 2023 Mar 14;29(1):34. doi: 10.1186/s10020-023-00628-3. Mol Med. 2023. PMID: 36918822 Free PMC article. Review.
-
Comparative analysis of the catalytic regulation of NEDD4-1 and WWP2 ubiquitin ligases.J Biol Chem. 2019 Nov 15;294(46):17421-17436. doi: 10.1074/jbc.RA119.009211. Epub 2019 Oct 2. J Biol Chem. 2019. PMID: 31578285 Free PMC article.
-
HECT E3 ubiquitin ligases - emerging insights into their biological roles and disease relevance.J Cell Sci. 2020 Apr 7;133(7):jcs228072. doi: 10.1242/jcs.228072. J Cell Sci. 2020. PMID: 32265230 Free PMC article. Review.
-
Oligomerization of the HECT ubiquitin ligase NEDD4-2/NEDD4L is essential for polyubiquitin chain assembly.J Biol Chem. 2018 Nov 23;293(47):18192-18206. doi: 10.1074/jbc.RA118.003716. Epub 2018 Oct 4. J Biol Chem. 2018. PMID: 30287686 Free PMC article.
Cited by
-
NEDD4 and NEDD4L: Ubiquitin Ligases Closely Related to Digestive Diseases.Biomolecules. 2024 May 13;14(5):577. doi: 10.3390/biom14050577. Biomolecules. 2024. PMID: 38785984 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

