Pathogenic human variant that dislocates GATA2 zinc fingers disrupts hematopoietic gene expression and signaling networks
- PMID: 36809258
- PMCID: PMC10065080
- DOI: 10.1172/JCI162685
Pathogenic human variant that dislocates GATA2 zinc fingers disrupts hematopoietic gene expression and signaling networks
Abstract
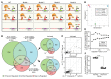
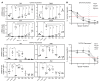
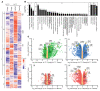
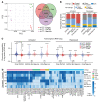
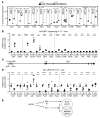
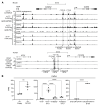
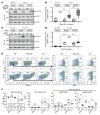
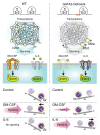
Although certain human genetic variants are conspicuously loss of function, decoding the impact of many variants is challenging. Previously, we described a patient with leukemia predisposition syndrome (GATA2 deficiency) with a germline GATA2 variant that inserts 9 amino acids between the 2 zinc fingers (9aa-Ins). Here, we conducted mechanistic analyses using genomic technologies and a genetic rescue system with Gata2 enhancer-mutant hematopoietic progenitor cells to compare how GATA2 and 9aa-Ins function genome-wide. Despite nuclear localization, 9aa-Ins was severely defective in occupying and remodeling chromatin and regulating transcription. Variation of the inter-zinc finger spacer length revealed that insertions were more deleterious to activation than repression. GATA2 deficiency generated a lineage-diverting gene expression program and a hematopoiesis-disrupting signaling network in progenitors with reduced granulocyte-macrophage colony-stimulating factor (GM-CSF) and elevated IL-6 signaling. As insufficient GM-CSF signaling caused pulmonary alveolar proteinosis and excessive IL-6 signaling promoted bone marrow failure and GATA2 deficiency patient phenotypes, these results provide insight into mechanisms underlying GATA2-linked pathologies.
Keywords: Genetic variation; Genetics; Hematology; Signal transduction; Transcription.
Conflict of interest statement
Figures



Similar articles
-
Loss of c-Kit and bone marrow failure upon conditional removal of the GATA-2 C-terminal zinc finger domain in adult mice.Eur J Haematol. 2016 Sep;97(3):261-70. doi: 10.1111/ejh.12719. Epub 2016 Jan 14. Eur J Haematol. 2016. PMID: 26660446 Free PMC article.
-
GATA2 deficiency and human hematopoietic development modeled using induced pluripotent stem cells.Blood Adv. 2018 Dec 11;2(23):3553-3565. doi: 10.1182/bloodadvances.2018017137. Blood Adv. 2018. PMID: 30538114 Free PMC article.
-
Breaking the spatial constraint between neighboring zinc fingers: a new germline mutation in GATA2 deficiency syndrome.Leukemia. 2021 Jan;35(1):264-268. doi: 10.1038/s41375-020-0820-2. Epub 2020 Apr 14. Leukemia. 2021. PMID: 32286542 No abstract available.
-
Human GATA2 mutations and hematologic disease: how many paths to pathogenesis?Blood Adv. 2020 Sep 22;4(18):4584-4592. doi: 10.1182/bloodadvances.2020002953. Blood Adv. 2020. PMID: 32960960 Free PMC article. Review.
-
Case Report: Missing zinc finger domains: hemophagocytic lymphohistiocytosis in a GATA2 deficiency patient triggered by non-tuberculous mycobacteriosis.Front Immunol. 2023 Aug 23;14:1191757. doi: 10.3389/fimmu.2023.1191757. eCollection 2023. Front Immunol. 2023. PMID: 37680631 Free PMC article. Review.
Cited by
-
Linking GATA2 to myeloid dysplasia and complex cytogenetics in adult myelodysplastic neoplasm and acute myeloid leukemia.Blood Adv. 2024 Jan 9;8(1):80-92. doi: 10.1182/bloodadvances.2023011554. Blood Adv. 2024. PMID: 38029365 Free PMC article.
-
Pathogenic GATA2 genetic variants utilize an obligate enhancer mechanism to distort a multilineage differentiation program.Proc Natl Acad Sci U S A. 2024 Mar 5;121(10):e2317147121. doi: 10.1073/pnas.2317147121. Epub 2024 Feb 29. Proc Natl Acad Sci U S A. 2024. PMID: 38422019 Free PMC article.
-
A gain-of-function p53 mutant synergizes with oncogenic NRAS to promote acute myeloid leukemia in mice.J Clin Invest. 2023 Dec 15;133(24):e173116. doi: 10.1172/JCI173116. J Clin Invest. 2023. PMID: 37847561 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

