This is a preprint.
Long COVID manifests with T cell dysregulation, inflammation, and an uncoordinated adaptive immune response to SARS-CoV-2
- PMID: 36798286
- PMCID: PMC9934605
- DOI: 10.1101/2023.02.09.527892
Long COVID manifests with T cell dysregulation, inflammation, and an uncoordinated adaptive immune response to SARS-CoV-2
Update in
-
Long COVID manifests with T cell dysregulation, inflammation and an uncoordinated adaptive immune response to SARS-CoV-2.Nat Immunol. 2024 Feb;25(2):218-225. doi: 10.1038/s41590-023-01724-6. Epub 2024 Jan 11. Nat Immunol. 2024. PMID: 38212464 Free PMC article.
Abstract
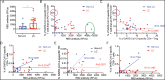
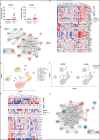
Long COVID (LC), a type of post-acute sequelae of SARS-CoV-2 infection (PASC), occurs after at least 10% of SARS-CoV-2 infections, yet its etiology remains poorly understood. Here, we used multiple "omics" assays (CyTOF, RNAseq/scRNAseq, Olink) and serology to deeply characterize both global and SARS-CoV-2-specific immunity from blood of individuals with clear LC and non-LC clinical trajectories, 8 months following infection and prior to receipt of any SARS-CoV-2 vaccine. Our analysis focused on deep phenotyping of T cells, which play important roles in immunity against SARS-CoV-2 yet may also contribute to COVID-19 pathogenesis. Our findings demonstrate that individuals with LC exhibit systemic inflammation and immune dysregulation. This is evidenced by global differences in T cell subset distribution in ways that imply ongoing immune responses, as well as by sex-specific perturbations in cytolytic subsets. Individuals with LC harbored increased frequencies of CD4+ T cells poised to migrate to inflamed tissues, and exhausted SARS-CoV-2-specific CD8+ T cells. They also harbored significantly higher levels of SARS-CoV-2 antibodies, and in contrast to non-LC individuals, exhibited a mis-coordination between their SARS-CoV-2-specific T and B cell responses. RNAseq/scRNAseq and Olink analyses similarly revealed immune dysregulatory mechanisms, along with non-immune associated perturbations, in individuals with LC. Collectively, our data suggest that proper crosstalk between the humoral and cellular arms of adaptive immunity has broken down in LC, and that this, perhaps in the context of persistent virus, leads to the immune dysregulation, inflammation, and clinical symptoms associated with this debilitating condition.
Conflict of interest statement
CONFLICTS OF INTERESTS MJP reports consulting fees from Gilead Sciences and AstraZeneca, outside the submitted work. SGD reports grants and/or personal fees from Gilead Sciences, Merck & Co., Viiv, AbbVie, Eli Lilly, ByroLogyx, and Enochian Biosciences, outside the submitted work. TJH receives grant support from Merck and consults for Roche. All other authors report no potential conflicts.
Figures

Similar articles
-
Long COVID manifests with T cell dysregulation, inflammation and an uncoordinated adaptive immune response to SARS-CoV-2.Nat Immunol. 2024 Feb;25(2):218-225. doi: 10.1038/s41590-023-01724-6. Epub 2024 Jan 11. Nat Immunol. 2024. PMID: 38212464 Free PMC article.
-
Systems analysis of innate and adaptive immunity in Long COVID.Semin Immunol. 2024 Mar;72:101873. doi: 10.1016/j.smim.2024.101873. Epub 2024 Mar 8. Semin Immunol. 2024. PMID: 38460395 Review.
-
Postacute sequelae and adaptive immune responses in people with HIV recovering from SARS-COV-2 infection.AIDS. 2022 Oct 1;36(12):F7-F16. doi: 10.1097/QAD.0000000000003338. Epub 2022 Jul 18. AIDS. 2022. PMID: 35866847 Free PMC article.
-
Long-term SARS-CoV-2-specific immune and inflammatory responses in individuals recovering from COVID-19 with and without post-acute symptoms.Cell Rep. 2021 Aug 10;36(6):109518. doi: 10.1016/j.celrep.2021.109518. Epub 2021 Aug 6. Cell Rep. 2021. PMID: 34358460 Free PMC article.
-
Immune mechanisms underlying COVID-19 pathology and post-acute sequelae of SARS-CoV-2 infection (PASC).Elife. 2023 May 26;12:e86014. doi: 10.7554/eLife.86014. Elife. 2023. PMID: 37233729 Free PMC article. Review.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
