The spatial distribution of GPCR and Gβγ activity across a cell dictates PIP3 dynamics
- PMID: 36797332
- PMCID: PMC9935898
- DOI: 10.1038/s41598-023-29639-0
The spatial distribution of GPCR and Gβγ activity across a cell dictates PIP3 dynamics
Abstract
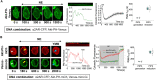
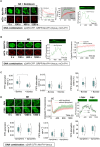
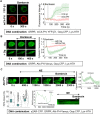
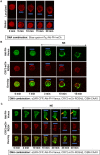
Phosphatidylinositol (3,4,5) trisphosphate (PIP3) is a plasma membrane-bound signaling phospholipid involved in many cellular signaling pathways that control crucial cellular processes and behaviors, including cytoskeleton remodeling, metabolism, chemotaxis, and apoptosis. Therefore, defective PIP3 signaling is implicated in various diseases, including cancer, diabetes, obesity, and cardiovascular diseases. Upon activation by G protein-coupled receptors (GPCRs) or receptor tyrosine kinases (RTKs), phosphoinositide-3-kinases (PI3Ks) phosphorylate phosphatidylinositol (4,5) bisphosphate (PIP2), generating PIP3. Though the mechanisms are unclear, PIP3 produced upon GPCR activation attenuates within minutes, indicating a tight temporal regulation. Our data show that subcellular redistributions of G proteins govern this PIP3 attenuation when GPCRs are activated globally, while localized GPCR activation induces sustained subcellular PIP3. Interestingly the observed PIP3 attenuation was Gγ subtype-dependent. Considering distinct cell-tissue-specific Gγ expression profiles, our findings not only demonstrate how the GPCR-induced PIP3 response is regulated depending on the GPCR activity gradient across a cell, but also show how diversely cells respond to spatial and temporal variability of external stimuli.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Statins Perturb Gβγ Signaling and Cell Behavior in a Gγ Subtype Dependent Manner.Mol Pharmacol. 2019 Apr;95(4):361-375. doi: 10.1124/mol.118.114710. Epub 2019 Feb 14. Mol Pharmacol. 2019. PMID: 30765461 Free PMC article.
-
PREX1 Protein Function Is Negatively Regulated Downstream of Receptor Tyrosine Kinase Activation by p21-activated Kinases (PAKs).J Biol Chem. 2016 Sep 16;291(38):20042-54. doi: 10.1074/jbc.M116.723882. Epub 2016 Aug 1. J Biol Chem. 2016. PMID: 27481946 Free PMC article.
-
Dissociation of the G protein βγ from the Gq-PLCβ complex partially attenuates PIP2 hydrolysis.J Biol Chem. 2021 Jan-Jun;296:100702. doi: 10.1016/j.jbc.2021.100702. Epub 2021 Apr 24. J Biol Chem. 2021. PMID: 33901492 Free PMC article.
-
Molecular regulation of PLCβ signaling.Methods Enzymol. 2023;682:17-52. doi: 10.1016/bs.mie.2023.01.001. Epub 2023 Feb 22. Methods Enzymol. 2023. PMID: 36948701 Review.
-
Phosphatidylinositol (3,4) bisphosphate-specific phosphatases and effector proteins: A distinct branch of PI3K signaling.Cell Signal. 2015 Sep;27(9):1789-98. doi: 10.1016/j.cellsig.2015.05.013. Epub 2015 May 27. Cell Signal. 2015. PMID: 26022180 Review.
Cited by
-
Aloe emodin promotes mucosal healing by modifying the differentiation fate of enteroendocrine cells via regulating cellular free fatty acid sensitivity.Acta Pharm Sin B. 2024 Sep;14(9):3964-3982. doi: 10.1016/j.apsb.2024.05.027. Epub 2024 May 31. Acta Pharm Sin B. 2024. PMID: 39309505 Free PMC article.
-
In-silico predicted mouse melanopsins with blue spectral shifts deliver efficient subcellular signaling.Cell Commun Signal. 2024 Aug 8;22(1):394. doi: 10.1186/s12964-024-01753-0. Cell Commun Signal. 2024. PMID: 39118111 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

