An in vitro approach to understand contribution of kidney cells to human urinary extracellular vesicles
- PMID: 36785873
- PMCID: PMC9925963
- DOI: 10.1002/jev2.12304
An in vitro approach to understand contribution of kidney cells to human urinary extracellular vesicles
Abstract
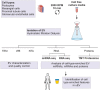
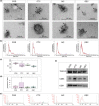
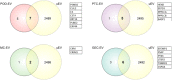
Extracellular vesicles (EV) are membranous particles secreted by all cells and found in body fluids. Established EV contents include a variety of RNA species, proteins, lipids and metabolites that are considered to reflect the physiological status of their parental cells. However, to date, little is known about cell-type enriched EV cargo in complex EV mixtures, especially in urine. To test whether EV secretion from distinct human kidney cells in culture differ and can recapitulate findings in normal urine, we comprehensively analysed EV components, (particularly miRNAs, long RNAs and protein) from conditionally immortalised human kidney cell lines (podocyte, glomerular endothelial, mesangial and proximal tubular cells) and compared to EV secreted in human urine. EV from cell culture media derived from immortalised kidney cells were isolated by hydrostatic filtration dialysis (HFD) and characterised by electron microscopy (EM), nanoparticle tracking analysis (NTA) and Western blotting (WB). RNA was isolated from EV and subjected to miRNA and RNA sequencing and proteins were profiled by tandem mass tag proteomics. Representative sets of EV miRNAs, RNAs and proteins were detected in each cell type and compared to human urinary EV isolates (uEV), EV cargo database, kidney biopsy bulk RNA sequencing and proteomics, and single-cell transcriptomics. This revealed that a high proportion of the in vitro EV signatures were also found in in vivo datasets. Thus, highlighting the robustness of our in vitro model and showing that this approach enables the dissection of cell type specific EV cargo in biofluids and the potential identification of cell-type specific EV biomarkers of kidney disease.
Keywords: RNA; exosomes; extracellular vesicles; glomerular endothelial cells; kidney; mesangial cells; miRNA; podocytes; proteomics; proximal tubule cells.
© 2023 The Authors. Journal of Extracellular Vesicles published by Wiley Periodicals, LLC on behalf of the International Society for Extracellular Vesicles.
Conflict of interest statement
The authors have no conflict of interest to declare.
Figures





Similar articles
-
Comparison of urinary extracellular vesicle isolation methods for transcriptomic biomarker research in diabetic kidney disease.J Extracell Vesicles. 2020 Dec;10(2):e12038. doi: 10.1002/jev2.12038. Epub 2021 Jan 7. J Extracell Vesicles. 2020. PMID: 33437407 Free PMC article.
-
Urinary extracellular vesicles: Assessment of pre-analytical variables and development of a quality control with focus on transcriptomic biomarker research.J Extracell Vesicles. 2021 Oct;10(12):e12158. doi: 10.1002/jev2.12158. J Extracell Vesicles. 2021. PMID: 34651466 Free PMC article.
-
Proteomic analysis of extracellular vesicles secreted by primary human epithelial endometrial cells reveals key proteins related to embryo implantation.Reprod Biol Endocrinol. 2022 Jan 3;20(1):3. doi: 10.1186/s12958-021-00879-x. Reprod Biol Endocrinol. 2022. PMID: 34980157 Free PMC article.
-
The protein and microRNA cargo of extracellular vesicles from parasitic helminths - current status and research priorities.Int J Parasitol. 2020 Aug;50(9):635-645. doi: 10.1016/j.ijpara.2020.04.010. Epub 2020 Jul 8. Int J Parasitol. 2020. PMID: 32652128 Review.
-
Circulating Y-RNAs in Extracellular Vesicles and Ribonucleoprotein Complexes; Implications for the Immune System.Front Immunol. 2019 Jan 15;9:3164. doi: 10.3389/fimmu.2018.03164. eCollection 2018. Front Immunol. 2019. PMID: 30697216 Free PMC article. Review.
Cited by
-
Isolation and Characterization of Cetacean Cell-Derived Extracellular Vesicles.Animals (Basel). 2023 Oct 24;13(21):3304. doi: 10.3390/ani13213304. Animals (Basel). 2023. PMID: 37958059 Free PMC article.
-
Exploring the role of urinary extracellular vesicles in kidney physiology, aging, and disease progression.Am J Physiol Cell Physiol. 2023 Dec 1;325(6):C1439-C1450. doi: 10.1152/ajpcell.00349.2023. Epub 2023 Oct 16. Am J Physiol Cell Physiol. 2023. PMID: 37842748 Free PMC article. Review.
-
Plant-Derived Exosome-Like Nanovesicles: Current Progress and Prospects.Int J Nanomedicine. 2023 Sep 5;18:4987-5009. doi: 10.2147/IJN.S420748. eCollection 2023. Int J Nanomedicine. 2023. PMID: 37693885 Free PMC article. Review.
-
Extracellular Vesicles: Investigating the Pathophysiology of Diabetes-Associated Hypertension and Diabetic Nephropathy.Biology (Basel). 2023 Aug 16;12(8):1138. doi: 10.3390/biology12081138. Biology (Basel). 2023. PMID: 37627022 Free PMC article. Review.
-
Plant-derived extracellular vesicles (PDEVs) in nanomedicine for human disease and therapeutic modalities.J Nanobiotechnology. 2023 Mar 29;21(1):114. doi: 10.1186/s12951-023-01858-7. J Nanobiotechnology. 2023. PMID: 36978093 Free PMC article. Review.
References
-
- Aguado‐Fraile, E. , Ramos, E. , Conde, E. , Rodríguez, M. , Martín‐Gómez, L. , Lietor, A. , Candela, Á. , Ponte, B. , Liaño, F. , & García‐Bermejo, M. L. (2015). A pilot study identifying a set of microRNAs as precise diagnostic biomarkers of acute kidney injury. PLoS ONE, 10(6), e0127175. 10.1371/journal.pone.0127175 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

