Regulation of autophagy and lipid accumulation under phosphate limitation in Rhodotorula toruloides
- PMID: 36777022
- PMCID: PMC9908577
- DOI: 10.3389/fmicb.2022.1046114
Regulation of autophagy and lipid accumulation under phosphate limitation in Rhodotorula toruloides
Abstract
Background: It is known that autophagy is essential for cell survival under stress conditions. Inorganic phosphate (Pi) is an essential nutrient for cell growth and Pi-limitation can trigger autophagy and lipid accumulation in oleaginous yeasts, yet protein (de)-phosphorylation and related signaling events in response to Pi limitation and the molecular basis linking Pi-limitation to autophagy and lipid accumulation remain elusive.
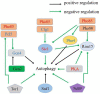
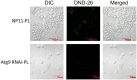
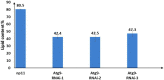
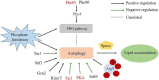
Results: Here, we compared the proteome and phosphoproteome of Rhodotorula toruloides CGMCC 2.1389 under Pi-limitation and Pi-repletion. In total, proteome analysis identified 3,556 proteins and the phosphoproteome analysis identified 1,649 phosphoproteins contained 5,659 phosphosites including 4,499 pSer, 978 pThr, and 182 pTyr. We found Pi-starvation-induced autophagy was regulated by autophagy-related proteins, but not the PHO pathway. When ATG9 was knocked down, the engineered strains produced significantly less lipids under Pi-limitation, suggesting that autophagy required Atg9 in R. toruloides and that was conducive to lipid accumulation.
Conclusion: Our results provide new insights into autophagy regulation under Pi-limitation and lipid accumulation in oleaginous yeast, which should be valuable to guide further mechanistic study of oleaginicity and genetic engineering for advanced lipid producing cell factory.
Keywords: Atg9; Rhodotorula toruloides; autophagy; lipid accumulation; phosphate limitation.
Copyright © 2023 Wang, Liu, Liu, Zhang, Jiao, Ye, Zhao and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Systems analysis of phosphate-limitation-induced lipid accumulation by the oleaginous yeast Rhodosporidium toruloides.Biotechnol Biofuels. 2018 May 25;11:148. doi: 10.1186/s13068-018-1134-8. eCollection 2018. Biotechnol Biofuels. 2018. PMID: 29849765 Free PMC article.
-
Nitrogen starvation causes lipid remodeling in Rhodotorula toruloides.Microb Cell Fact. 2024 May 17;23(1):141. doi: 10.1186/s12934-024-02414-0. Microb Cell Fact. 2024. PMID: 38760782 Free PMC article.
-
Reduction of lipid-accumulation of oleaginous yeast Rhodosporidium toruloides through CRISPR/Cas9-mediated inactivation of lipid droplet structural proteins.FEMS Microbiol Lett. 2021 Sep 1;368(16):fnab111. doi: 10.1093/femsle/fnab111. FEMS Microbiol Lett. 2021. PMID: 34410383
-
Rhodotorula toruloides: an ideal microbial cell factory to produce oleochemicals, carotenoids, and other products.World J Microbiol Biotechnol. 2021 Dec 7;38(1):13. doi: 10.1007/s11274-021-03201-4. World J Microbiol Biotechnol. 2021. PMID: 34873661 Review.
-
Development and Perspective of Rhodotorula toruloides as an Efficient Cell Factory.J Agric Food Chem. 2023 Feb 1;71(4):1802-1819. doi: 10.1021/acs.jafc.2c07361. Epub 2023 Jan 23. J Agric Food Chem. 2023. PMID: 36688927 Review.
Cited by
-
Corn stover variability drives differences in bisabolene production by engineered Rhodotorula toruloides.J Ind Microbiol Biotechnol. 2024 Jan 9;51:kuae034. doi: 10.1093/jimb/kuae034. J Ind Microbiol Biotechnol. 2024. PMID: 39317673 Free PMC article.
-
Decreased mRNA expression of NR1H3 and ABCA1 in pulmonary tuberculosis patients from population of Punjab, India.Mol Biol Rep. 2024 May 13;51(1):657. doi: 10.1007/s11033-024-09589-0. Mol Biol Rep. 2024. PMID: 38740636
References
-
- Ballif B. A., Roux P. P., Gerber S. A., Mac Keigan J. P., Blenis J., Gygi S. P. (2005). Quantitative phosphorylation profiling of the ERK/p 90 ribosomal S6 kinase-signaling cassette and its targets, the tuberous sclerosis tumor suppressors. Proc. Natl. Acad. Sci. U. S. A. 102, 667–672. doi: 10.1073/pnas.0409143102, PMID: - DOI - PMC - PubMed
-
- Budovskaya Y. V., Stephan J. S., Reggiori F., Klionsky D. J., Herman P. K. (2004). The Ras/cAMP-dependent protein kinase signaling pathway regulates an early step of the autophagy process in Saccharomyces cerevisiae. J. Biol. Chem. 279, 20663–20671. doi: 10.1074/jbc.M400272200, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

