Circulating Chromosome Conformation Signatures Significantly Enhance PSA Positive Predicting Value and Overall Accuracy for Prostate Cancer Detection
- PMID: 36765779
- PMCID: PMC9913359
- DOI: 10.3390/cancers15030821
Circulating Chromosome Conformation Signatures Significantly Enhance PSA Positive Predicting Value and Overall Accuracy for Prostate Cancer Detection
Abstract
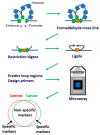
Background: Prostate cancer (PCa) has a high lifetime prevalence (one out of six men), but currently there is no widely accepted screening programme. Widely used prostate specific antigen (PSA) test at cut-off of 3.0 ng/mL does not have sufficient accuracy for detection of any prostate cancer, resulting in numerous unnecessary prostate biopsies in men with benign disease and false reassurance in some men with PCa. We have recently identified circulating chromosome conformation signatures (CCSs, Episwitch® PCa test) allowing PCa detection and risk stratification in line with standards of clinical PCa staging. The purpose of this study was to determine whether combining the Episwitch PCa test with the PSA test will increase its diagnostic accuracy.
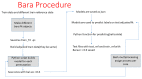
Methods: n = 109 whole blood samples of men enrolled in the PROSTAGRAM screening pilot study and n = 38 samples of patients with established PCa diagnosis and cancer-negative controls from Imperial College NHS Trust were used. Samples were tested for PSA, and the presence of CCSs in the loci encoding for of DAPK1, HSD3B2, SRD5A3, MMP1, and miRNA98 associated with high-risk PCa identified in our previous work.
Results: PSA > 3 ng/mL alone showed a low positive predicted value (PPV) of 0.14 and a high negative predicted value (NPV) of 0.93. EpiSwitch alone showed a PPV of 0.91 and a NPV of 0.32. Combining PSA and Episwitch tests has significantly increased the PPV to 0.81 although reducing the NPV to 0.78. Furthermore, integrating PSA, as a continuous variable (rather than a dichotomised 3 ng/mL cut-off), with EpiSwitch in a new multivariant stratification model, Prostate Screening EpiSwitch (PSE) test, has yielded a remarkable combined PPV of 0.92 and NPV of 0.94 when tested on the independent prospective cohort.
Conclusions: Our results demonstrate that combining the standard PSA readout with circulating chromosome conformations (PSE test) allows for significantly enhanced PSA PPV and overall accuracy for PCa detection. The PSE test is accurate, rapid, minimally invasive, and inexpensive, suggesting significant screening diagnostic potential to minimise unnecessary referrals for expensive and invasive MRI and/or biopsy testing. Further extended prospective blinded validation of the new combined signature in a screening cohort with low cancer prevalence would be the recommended step for PSE adoption in PCa screening.
Keywords: PSA; blood test; diagnosis; epigenetics; nucleome; prostate cancer; screening.
Conflict of interest statement
Ewan Hunter, Matthew Salter, Mehrnoush Dezfouli, Ryan Powel, Jayne Green, Tarun Naithani, Christina Koutsothanasi, and Alexandre Akoulitchev are employees of Oxford BioDynamics. A. Akoulitchev is a company director. Oxford BioDynamics holds patents on the EpiSwitch technology. The remaining authors have no conflict of interest.
Figures


Similar articles
-
Chromatin conformation changes in peripheral blood can detect prostate cancer and stratify disease risk groups.J Transl Med. 2021 Jan 28;19(1):46. doi: 10.1186/s12967-021-02710-y. J Transl Med. 2021. PMID: 33509203 Free PMC article.
-
More advantages in detecting bone and soft tissue metastases from prostate cancer using 18F-PSMA PET/CT.Hell J Nucl Med. 2019 Jan-Apr;22(1):6-9. doi: 10.1967/s002449910952. Epub 2019 Mar 7. Hell J Nucl Med. 2019. PMID: 30843003
-
Predicting the Diagnosis of Prostate Cancer with a Novel Blood-Based Biomarker: Comparison of Its Performance with Prostate-Specific Antigen.Cancers (Basel). 2024 Jul 23;16(15):2619. doi: 10.3390/cancers16152619. Cancers (Basel). 2024. PMID: 39123347 Free PMC article.
-
Multiparametric MRI in detection and staging of prostate cancer.Dan Med J. 2017 Feb;64(2):B5327. Dan Med J. 2017. PMID: 28157066 Review.
-
Can Negative Prostate-specific Membrane Antigen Positron Emission Tomography/Computed Tomography Avoid the Need for Pelvic Lymph Node Dissection in Newly Diagnosed Prostate Cancer Patients? A Systematic Review and Meta-analysis with Backup Histology as Reference Standard.Eur Urol Oncol. 2022 Feb;5(1):1-17. doi: 10.1016/j.euo.2021.08.001. Epub 2021 Sep 17. Eur Urol Oncol. 2022. PMID: 34538770 Review.
Cited by
-
Development and Validation of Blood-Based Predictive Biomarkers for Response to PD-1/PD-L1 Checkpoint Inhibitors: Evidence of a Universal Systemic Core of 3D Immunogenetic Profiling across Multiple Oncological Indications.Cancers (Basel). 2023 May 10;15(10):2696. doi: 10.3390/cancers15102696. Cancers (Basel). 2023. PMID: 37345033 Free PMC article.
-
Cost Analysis of Prostate Cancer Care Using a Biomarker-enhanced Diagnostic Strategy with Stockholm3.Eur Urol Open Sci. 2024 Jun 25;66:26-32. doi: 10.1016/j.euros.2024.05.010. eCollection 2024 Aug. Eur Urol Open Sci. 2024. PMID: 39027655 Free PMC article.
References
-
- ACS American Cancer Society. 2020. [(accessed on 1 October 2022)]. Available online: http://www.cancer.org/cancer/prostatecancer/overviewguide/prostate-cance....
-
- CRUK Cancer Research UK. 2020. [(accessed on 1 October 2022)]. Available online: http://www.cancerresearchuk.org/home/
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

