Detection of coronaviruses in insectivorous bats of Fore-Caucasus, 2021
- PMID: 36759670
- PMCID: PMC9909659
- DOI: 10.1038/s41598-023-29099-6
Detection of coronaviruses in insectivorous bats of Fore-Caucasus, 2021
Abstract
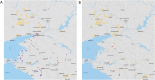
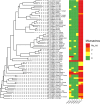
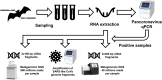
Coronaviruses (CoVs) pose a huge threat to public health as emerging viruses. Bat-borne CoVs are especially unpredictable in their evolution due to some unique features of bat physiology boosting the rate of mutations in CoVs, which is already high by itself compared to other viruses. Among bats, a meta-analysis of overall CoVs epizootiology identified a nucleic acid observed prevalence of 9.8% (95% CI 8.7-10.9%). The main objectives of our study were to conduct a qPCR screening of CoVs' prevalence in the insectivorous bat population of Fore-Caucasus and perform their characterization based on the metagenomic NGS of samples with detected CoV RNA. According to the qPCR screening, CoV RNA was detected in 5 samples, resulting in a 3.33% (95% CI 1.1-7.6%) prevalence of CoVs in bats from these studied locations. BetaCoVs reads were identified in raw metagenomic NGS data, however, detailed characterization was not possible due to relatively low RNA concentration in samples. Our results correspond to other studies, although a lower prevalence in qPCR studies was observed compared to other regions and countries. Further studies should require deeper metagenomic NGS investigation, as a supplementary method, which will allow detailed CoV characterization.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Coronavirus and paramyxovirus in bats from Northwest Italy.BMC Vet Res. 2017 Dec 22;13(1):396. doi: 10.1186/s12917-017-1307-x. BMC Vet Res. 2017. PMID: 29273042 Free PMC article.
-
Surveillance of Bat Coronaviruses in Kenya Identifies Relatives of Human Coronaviruses NL63 and 229E and Their Recombination History.J Virol. 2017 Feb 14;91(5):e01953-16. doi: 10.1128/JVI.01953-16. Print 2017 Mar 1. J Virol. 2017. PMID: 28077633 Free PMC article.
-
Global Epidemiology of Bat Coronaviruses.Viruses. 2019 Feb 20;11(2):174. doi: 10.3390/v11020174. Viruses. 2019. PMID: 30791586 Free PMC article. Review.
-
Neotropical Bats from Costa Rica harbour Diverse Coronaviruses.Zoonoses Public Health. 2015 Nov;62(7):501-5. doi: 10.1111/zph.12181. Epub 2015 Feb 4. Zoonoses Public Health. 2015. PMID: 25653111 Free PMC article.
-
Ecology, evolution and classification of bat coronaviruses in the aftermath of SARS.Antiviral Res. 2014 Jan;101:45-56. doi: 10.1016/j.antiviral.2013.10.013. Epub 2013 Oct 31. Antiviral Res. 2014. PMID: 24184128 Free PMC article. Review.
Cited by
-
Gut Microbiota Composition of Insectivorous Synanthropic and Fructivorous Zoo Bats: A Direct Metagenomic Comparison.Int J Mol Sci. 2023 Dec 9;24(24):17301. doi: 10.3390/ijms242417301. Int J Mol Sci. 2023. PMID: 38139130 Free PMC article.
-
Cultivable Gut Microbiota in Synanthropic Bats: Shifts of Its Composition and Diversity Associated with Hibernation.Animals (Basel). 2023 Nov 26;13(23):3658. doi: 10.3390/ani13233658. Animals (Basel). 2023. PMID: 38067008 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

