Allele-specific silencing as therapy for familial amyotrophic lateral sclerosis caused by the p.G376D TARDBP mutation
- PMID: 36751500
- PMCID: PMC9897181
- DOI: 10.1093/braincomms/fcac315
Allele-specific silencing as therapy for familial amyotrophic lateral sclerosis caused by the p.G376D TARDBP mutation
Abstract
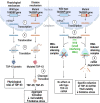
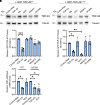
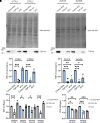
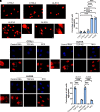
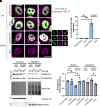
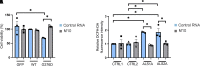
Amyotrophic lateral sclerosis is a neurodegenerative disease characterized by the degeneration of motor neurons. There is no treatment for this disease that affects the ability to move, eat, speak and finally breathe, causing death. In an Italian family, a heterozygous pathogenic missense variant has been previously discovered in Exon 6 of the gene TARDBP encoding the TAR DNA-binding protein 43 protein. Here, we developed a potential therapeutic tool based on allele-specific small interfering RNAs for familial amyotrophic lateral sclerosis with the heterozygous missense mutation c.1127G>A. We designed a small interfering RNA that was able to diminish specifically the expression of the exogenous Green Fluorescent Protein (TAR DNA-binding protein 43G376D mutant protein) in HEK-293T cells but not that of the Green Fluorescent Protein (TAR DNA-binding protein 43 wild-type). Similarly, this small interfering RNA silenced the mutated allele in fibroblasts derived from patients with amyotrophic lateral sclerosis but did not silence the wild-type gene in control fibroblasts. In addition, we established that silencing the mutated allele was able to strongly reduce the pathological cellular phenotypes induced by TAR DNA-binding protein 43G376D expression, such as the presence of cytoplasmic aggregates. Thus, we have identified a small interfering RNA that could be used to silence specifically the mutated allele to try a targeted therapy for patients carrying the p.G376D TAR DNA-binding protein 43 mutation.
Keywords: ALS; TDP-43; allele-specific silencing; antisense oligonucleotides; siRNA therapy.
© The Author(s) 2022. Published by Oxford University Press on behalf of the Guarantors of Brain.
Figures
Similar articles
-
Novel mutations in TARDBP (TDP-43) in patients with familial amyotrophic lateral sclerosis.PLoS Genet. 2008 Sep 19;4(9):e1000193. doi: 10.1371/journal.pgen.1000193. PLoS Genet. 2008. PMID: 18802454 Free PMC article.
-
Production of CSSi013-A (9360) iPSC line from an asymptomatic subject carrying an heterozygous mutation in TDP-43 protein.Stem Cell Res. 2022 Aug;63:102835. doi: 10.1016/j.scr.2022.102835. Epub 2022 Jun 6. Stem Cell Res. 2022. PMID: 35714448
-
High frequency of TARDBP gene mutations in Italian patients with amyotrophic lateral sclerosis.Hum Mutat. 2009 Apr;30(4):688-94. doi: 10.1002/humu.20950. Hum Mutat. 2009. PMID: 19224587
-
Mutations in TDP-43 link glycine-rich domain functions to amyotrophic lateral sclerosis.Hum Mol Genet. 2009 Oct 15;18(R2):R156-62. doi: 10.1093/hmg/ddp303. Hum Mol Genet. 2009. PMID: 19808791 Free PMC article. Review.
-
TARDBP and FUS mutations associated with amyotrophic lateral sclerosis: summary and update.Hum Mutat. 2013 Jun;34(6):812-26. doi: 10.1002/humu.22319. Epub 2013 Apr 29. Hum Mutat. 2013. PMID: 23559573 Review.
Cited by
-
The new missense G376V-TDP-43 variant induces late-onset distal myopathy but not amyotrophic lateral sclerosis.Brain. 2024 May 3;147(5):1768-1783. doi: 10.1093/brain/awad410. Brain. 2024. PMID: 38079474 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Miscellaneous
