Hi-C analysis of genomic contacts revealed karyotype abnormalities in chicken HD3 cell line
- PMID: 36750787
- PMCID: PMC9906895
- DOI: 10.1186/s12864-023-09158-y
Hi-C analysis of genomic contacts revealed karyotype abnormalities in chicken HD3 cell line
Abstract
Background: Karyotype abnormalities are frequent in immortalized continuous cell lines either transformed or derived from primary tumors. Chromosomal rearrangements can cause dramatic changes in gene expression and affect cellular phenotype and behavior during in vitro culture. Structural variations of chromosomes in many continuous mammalian cell lines are well documented, but chromosome aberrations in cell lines from other vertebrate models often remain understudied. The chicken LSCC-HD3 cell line (HD3), generated from erythroid precursors, was used as an avian model for erythroid differentiation and lineage-specific gene expression. However, karyotype abnormalities in the HD3 cell line were not assessed. In the present study, we applied high-throughput chromosome conformation capture to analyze 3D genome organization and to detect chromosome rearrangements in the HD3 cell line.
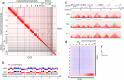
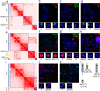
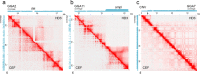
Results: We obtained Hi-C maps of genomic interactions for the HD3 cell line and compared A/B compartments and topologically associating domains between HD3 and several other cell types. By analysis of contact patterns in the Hi-C maps of HD3 cells, we identified more than 25 interchromosomal translocations of regions ≥ 200 kb on both micro- and macrochromosomes. We classified most of the observed translocations as unbalanced, leading to the formation of heteromorphic chromosomes. In many cases of microchromosome rearrangements, an entire microchromosome together with other macro- and microchromosomes participated in the emergence of a derivative chromosome, resembling "chromosomal fusions'' between acrocentric microchromosomes. Intrachromosomal inversions, deletions and duplications were also detected in HD3 cells. Several of the identified simple and complex chromosomal rearrangements, such as between GGA2 and GGA1qter; GGA5, GGA4p and GGA7p; GGA4q, GGA6 and GGA19; and duplication of the sex chromosome GGAW, were confirmed by FISH.
Conclusions: In the erythroid progenitor HD3 cell line, in contrast to mature and immature erythrocytes, the genome is organized into distinct topologically associating domains. The HD3 cell line has a severely rearranged karyotype with most of the chromosomes engaged in translocations and can be used in studies of genome structure-function relationships. Hi-C proved to be a reliable tool for simultaneous assessment of the spatial genome organization and chromosomal aberrations in karyotypes of birds with a large number of microchromosomes.
Keywords: A/B compartments; Avian erythroid differentiation; Avian karyotype; Chicken cells; Chromosome rearrangement; Chromosome translocation; Genome architecture; HD3; Hi-C; Topologically associating domains.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Similar articles
-
Chromosomal Analysis in Crotophaga ani (Aves, Cuculiformes) Reveals Extensive Genomic Reorganization and an Unusual Z-Autosome Robertsonian Translocation.Cells. 2020 Dec 22;10(1):4. doi: 10.3390/cells10010004. Cells. 2020. PMID: 33375072 Free PMC article.
-
Interspecies Chromosome Mapping in Caprimulgiformes, Piciformes, Suliformes, and Trogoniformes (Aves): Cytogenomic Insight into Microchromosome Organization and Karyotype Evolution in Birds.Cells. 2021 Apr 7;10(4):826. doi: 10.3390/cells10040826. Cells. 2021. PMID: 33916942 Free PMC article.
-
Comparative chromosome maps between the stone curlew and three ciconiiform species (the grey heron, little egret and crested ibis).BMC Ecol Evol. 2022 Mar 3;22(1):23. doi: 10.1186/s12862-022-01979-x. BMC Ecol Evol. 2022. PMID: 35240987 Free PMC article.
-
Time lapse: A glimpse into prehistoric genomics.Eur J Med Genet. 2020 Feb;63(2):103640. doi: 10.1016/j.ejmg.2019.03.004. Epub 2019 Mar 25. Eur J Med Genet. 2020. PMID: 30922926 Free PMC article. Review.
-
[Micro vs. macro: structural-functional organization of avian micro- and macrochromosomes].Genetika. 1996 May;32(5):597-608. Genetika. 1996. PMID: 8755033 Review. Russian.
Cited by
-
Expanding the list of sequence-agnostic enzymes for chromatin conformation capture assays with S1 nuclease.Epigenetics Chromatin. 2023 Dec 11;16(1):48. doi: 10.1186/s13072-023-00524-4. Epigenetics Chromatin. 2023. PMID: 38072950 Free PMC article.
-
Assignment of the somatic A/B compartments to chromatin domains in giant transcriptionally active lampbrush chromosomes.Epigenetics Chromatin. 2023 Jun 15;16(1):24. doi: 10.1186/s13072-023-00499-2. Epigenetics Chromatin. 2023. PMID: 37322523 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

